Human endogenous retrovirus ERVK3-1 characterizes a metabolically active and immunosuppressive subtype of liver cancer
- PMID: 41034947
- PMCID: PMC12486636
- DOI: 10.1186/s12985-025-02928-y
Human endogenous retrovirus ERVK3-1 characterizes a metabolically active and immunosuppressive subtype of liver cancer
Abstract
Background: Human endogenous retroviruses (HERVs), particularly the HERV-K family, are increasingly recognized for their roles in cancer biology, yet the function of ERVK3-1 (HERVK_3p21.31) in liver hepatocellular carcinoma (LIHC) remains largely unexplored.
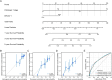
Methods: We analyzed transcriptomic data from the TCGA-LIHC cohort to identify differentially expressed genes (DEGs) between high and low ERVK3-1 expression groups, followed by functional enrichment analyses (GO, KEGG, GSEA), protein-protein interaction (PPI) network construction, and hub gene identification. The immunological relevance of ERVK3-1 was assessed through TIP immune cycle analysis, single-cell RNA sequencing datasets, and correlation with immune checkpoint expression. Immunotherapy responsiveness was evaluated using TIDE and TCIA databases.
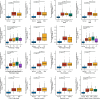
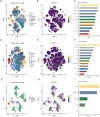
Results: High ERVK3-1 expression was associated with enrichment in metabolic and oxidative stress-related pathways, while low expression correlated with cell cycle and DNA replication. PPI analysis revealed mitosis-related hub genes (e.g., CCNB1, CDK1). ERVK3-1 expression promoted early immune cell recruitment but was inversely correlated with later stages of the cancer immunity cycle, including immune infiltration and T-cell killing. Single-cell data showed high ERVK3-1 expression in immunosuppressive subsets, alongside positive associations with inhibitory immune checkpoints (e.g., PD-1, CTLA-4, TIM-3). High ERVK3-1 expression also correlated with greater immune evasion and reduced immunotherapy responsiveness.
Conclusions: ERVK3-1 plays a multifaceted role in LIHC progression, contributing to metabolic reprogramming, immune suppression, and resistance to immunotherapy. These findings highlight ERVK3-1 as a potential prognostic biomarker and therapeutic target in liver cancer.
Keywords: ERVK3-1; Immunotherapy; Liver hepatocellular carcinoma; T-cell exhaustion; Tumor microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures






References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2021;71(3):209–49. - PubMed
-
- Ren Z, Xu J, Bai Y, Xu A, Cang S, Du C, et al. Sintilimab plus a bevacizumab biosimilar (IBI305) versus Sorafenib in unresectable hepatocellular carcinoma (ORIENT-32): a randomised, open-label, phase 2–3 study. Lancet Oncol. 2021;22(7):977–90. - PubMed
-
- Eggermont AMM, Blank CU, Mandala M, Long GV, Atkinson V, Dalle S, et al. Adjuvant pembrolizumab versus placebo in resected stage III melanoma. N Engl J Med. 2018;378(19):1789–801. - PubMed
-
- Cheng AL, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY, et al. Updated efficacy and safety data from IMbrave150: Atezolizumab plus bevacizumab vs. sorafenib for unresectable hepatocellular carcinoma. J Hepatol. 2022;76(4):862–73. - PubMed
-
- Ringelhan M, Pfister D, O’Connor T, Pikarsky E, Heikenwalder M. The immunology of hepatocellular carcinoma. Nat Immunol. 2018;19(3):222–32. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

