Immunoproteasome Inhibition Positively Impacts the Gut-Muscle Axis in Duchenne Muscular Dystrophy
- PMID: 41035224
- PMCID: PMC12489020
- DOI: 10.1002/jcsm.70054
Immunoproteasome Inhibition Positively Impacts the Gut-Muscle Axis in Duchenne Muscular Dystrophy
Abstract
Background: Duchenne Muscular Dystrophy (DMD) features immune-muscle crosstalk, where muscle fibre degeneration enhances pro-inflammatory macrophage infiltration, worsening inflammation and impairing regeneration.
Methods: We investigated the impact of immunoproteasome (IP) inhibition on the gut-muscle axis in mdx mice, a well-established model of DMD. We employed microbiota perturbation models, including broad-spectrum antibiotic treatment (ABX) and faecal microbiota transplantation (FMT) from IP-inhibited mdx mice. IP inhibition effects were assessed by analysing gut microbiota composition, intestinal inflammation, muscle integrity and associated metabolic and inflammatory pathways.
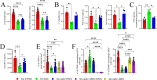
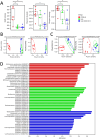
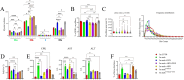
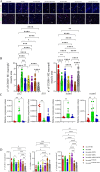
Results: IP inhibitor ONX-0914 significantly impacted the intestinal inflammatory microenvironment and gut microbiota of mdx mice. ONX-0914 treatment increased gastrointestinal transit (increased wet/dry faecal weights, p = 0.0486 and p = 0.0112, respectively) and partially restored intestinal barrier integrity (reduced FITC-dextran leakage, p = 0.0449). JAM-A was significantly upregulated (p < 0.0001). Colonic CD206+ M2 macrophages increased, while CD68 + M1 cells partially decreased. ONX-0914 downregulated IP isoforms in macrophages (PSMB8: p = 0.0022; PSMB9: p = 0.0186) as well as FOXO-1 (p = 0.0380) and TNF-α (p = 0.0487). Antibiotic-induced microbiota depletion abrogated these effects. Metagenomic analysis revealed significant differences in microbiota composition between C57Bl controls and mdx mice (PERMANOVA p < 0.001), with ONX-0914 inducing enrichment of stachyose degradation pathways. Metabolomic analysis showed enrichment of bacterial metabolites, fatty acid and sugar metabolism pathways, with increased glutathione, galactose, glycerol, glyceraldehyde and TCA cycle intermediates. ONX-0914 improved mitochondrial activity in skeletal muscle, as increased expression of ETC complexes (mdx vs. mdx+ONX: Complex II, p = 0.0338; Complex IV, p = 0.0023) and TCA enzymes (mdx vs. FTMmdx+ONX: IDH p = 0.0258; FH p = 0.0366). This led to a shift towards oxidative muscle fibres and improved muscle morphology (increased fibre size, p < 0.0001 mdx vs. mdx+ONX and mdx vs. FTMmdx+ONX). Muscle performance was enhanced with reduced CPK levels (p = 0.0015 mdx vs. mdx+ONX) and fibrosis (decreased TGFβ: mdx vs. mdx+ONX, p = 0.0248; mdx vs. FTMmdx+ONX, p = 0.0279). ONX-0914 reduced CD68+ (mdx vs. mdx+ONX, p = 0.0024; mdx vs. FTMmdx+ONX, p < 0.0001) and increased CD206+ (mdx vs. FTMmdx+ONX: p = 0.0083) macrophages in muscle, downregulated inflammatory genes (mdx vs. mdx+ONX: ccl2 p = 0.0327, vcam-1p = 0.0378) and reduced pro-inflammatory proteins (MCP1, mdx vs. mdx+ONX, p = 0.0442). Inflammatory cytokines and endothelial vessel density in ONX-0914 treated mdx were restored to wild type mice. These data demonstrate that ONX-0914 enhances muscle function through microbiota-dependent mechanisms.
Conclusions: Our study advances the understanding of the role of dysbiosis in DMD disease and identifies IP inhibition as a potential therapeutic strategy to modulate the dystrophic gut-muscle axis, offering new perspectives for microbiota-targeted therapies.
Keywords: Duchenne muscular dystrophy; immunoproteasome; macrophages; muscle metabolism.
© 2025 The Author(s). Journal of Cachexia, Sarcopenia and Muscle published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Briguet A., Courdier‐Fruh I., Foster M., Meier T., and Magyar J. P., “Histological Parameters for the Quantitative Assessment of Muscular Dystrophy in the Mdx‐Mouse,” Neuromuscular Disorders 14 (2004): 675–682. - PubMed
-
- Chazaud B., “Inflammation and Skeletal Muscle Regeneration: Leave It to the Macrophages!,” Trends in Immunology 41 (2020): 481–492. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

