Viral community diversity in the rhizosphere of the foundation salt marsh plant Spartina alterniflora
- PMID: 41036845
- PMCID: PMC12570469
- DOI: 10.1128/msphere.00234-25
Viral community diversity in the rhizosphere of the foundation salt marsh plant Spartina alterniflora
Abstract
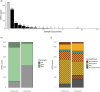
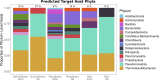
Viruses of microorganisms impact microbial population dynamics, community structure, nutrient cycling, gene transfer, and genomic innovation. In wetlands, root-associated microbial communities mediate key biogeochemical processes important for plants involved in ecosystem maintenance. Nonetheless, the presence and role of microbial viruses in salt marshes remain poorly understood. In this study, we analyzed 24 metagenomes retrieved from the root zone of Spartina alterniflora, a foundation plant in salt marshes of the eastern and Gulf coasts of the U.S. The samples span three plant compartments-bulk sediment, rhizosphere, and root-and two cordgrass plant phenotypes: short and tall. We observed differentiation between phenotypes and increased similarity in viral communities between the root and rhizosphere, indicating that plant compartment and phenotype shape viral community composition. The majority of viral populations characterized are novel at the genus level, with a subset predicted to target microorganisms known to carry out key biogeochemical functions. The findings contribute to ongoing efforts to understand plant-associated viral diversity and community composition and to identify potential targets for exploring viral modulation of microbially mediated ecosystem functioning in intertidal wetlands.IMPORTANCESalt marshes are vital coastal ecosystems. Microbes in these environments drive nutrient cycling and support plant health, with Spartina alterniflora serving as a foundation species. This study explores viral communities associated with S. alterniflora, revealing how plant compartments and phenotypes shape viral composition. The discovery of numerous novel viruses, some potentially influencing microbes involved in key biogeochemical processes, highlights their ecological significance. Given the increasing pressures on coastal ecosystems, understanding virus-microbe-plant interactions is essential for predicting and managing ecosystem responses to environmental change.
Keywords: bacteriophages; metagenomics; microbial ecology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Mitsch W, Gosselink J. 1993. Wetlands. 2nd ed. John Wiley and Sons, New York.
-
- Yang R-M, Guo W-W. 2018. Exotic Spartina alterniflora enhances the soil functions of a coastal ecosystem. Soil Sci Soc Am J 82:901–909. doi: 10.2136/sssaj2017.12.0411 - DOI
-
- Rolando JL, Kolton M, Song T, Liu Y, Pinamang P, Conrad R, Morris JT, Konstantinidis KT, Kostka JE. 2024. Sulfur oxidation and reduction are coupled to nitrogen fixation in the roots of the salt marsh foundation plant Spartina alterniflora. Nat Commun 15:3607. doi: 10.1038/s41467-024-47646-1 - DOI - PMC - PubMed
-
- Wilhelm SW, Suttle CA. 1999. Viruses and nutrient cycles in the sea. Bioscience 49:781–788. doi: 10.2307/1313569 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
