Single-Tube Capture, Concentration, and Genomic Extraction of a Human Norovirus Surrogate Using Magnetic Ionic Liquids
- PMID: 41038607
- PMCID: PMC12529475
- DOI: 10.1021/acs.analchem.5c03750
Single-Tube Capture, Concentration, and Genomic Extraction of a Human Norovirus Surrogate Using Magnetic Ionic Liquids
Abstract
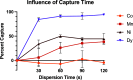
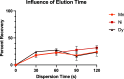
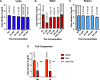
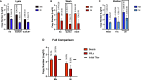
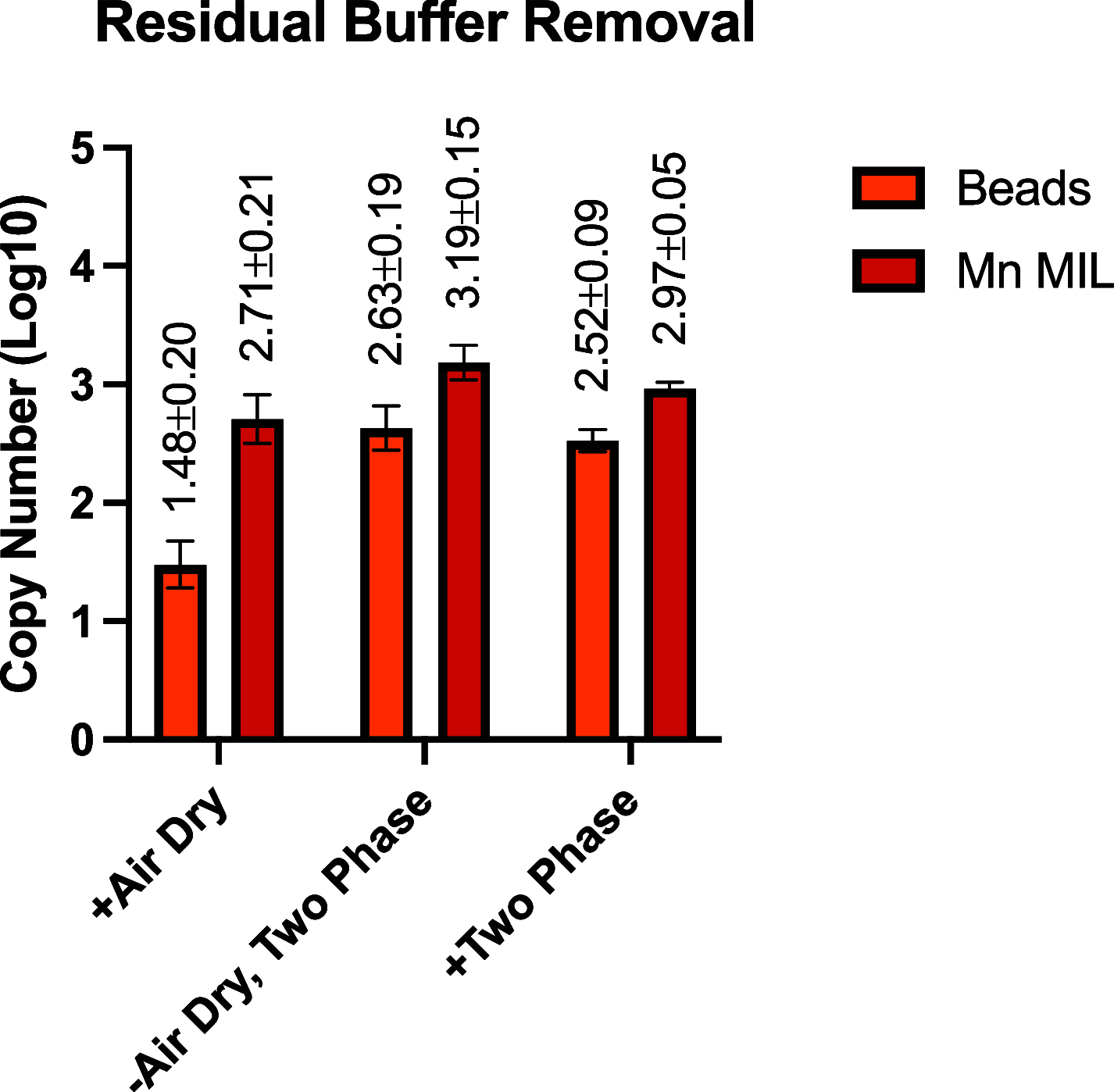
Capture and concentration of viruses from contaminated foods are crucial for sensitive detection. Magnetic ionic liquids (MILs) are ideal capture reagents for portable applications since they require minimal equipment and no cold storage. However, a similarly portable genomic extraction method is also needed for use with portable nucleic-acid-based detection methods. Since MILs effectively bind both intact virus and viral RNA, they can potentially be used as a binding substrate for both viral capture and genomic extraction. To evaluate this, a protocol was developed for single-tube capture, concentration, and genomic extraction based on a commercial method for magnetic bead-based RNA extraction that utilized MILs as the binding substrate. Two of the MIL formulations showed a recovery efficiency comparable to that of commercial magnetic beads and were capable of target enrichment when lysis, wash, and elution buffers from a commercial kit were used. These commercial buffers were proprietary and optimized for use with magnetic silica beads; therefore, next, various combinations of known lysis, wash, and elution buffers were tested with both magnetic silica beads and MILs until RNA recovery was comparable to that achieved with the commercial reagents. Though further research is needed, this work represents the first reported instance of single-tube capture, concentration, and genomic extraction of a microbial target using MILs as a universal substrate capable of mediating all three actions and a step toward fully portable microbial detection.
Figures
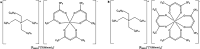


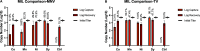
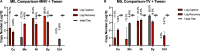
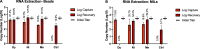
References
-
- Ahmed S. M., Hall A. J., Robinson A. E., Verhoef L., Premkumar P., Parashar U. D., Koopmans M., Lopman B. A.. Global Prevalence of Norovirus in Cases of Gastroenteritis: A Systematic Review and Meta-Analysis. Lancet Infect. Dis. 2014;14(8):725–730. doi: 10.1016/S1473-3099(14)70767-4. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

