Platelet activation plays a pro-inflammatory role in myasthenia gravis
- PMID: 41038808
- PMCID: PMC12491536
- DOI: 10.1038/s41467-025-63750-2
Platelet activation plays a pro-inflammatory role in myasthenia gravis
Abstract
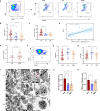
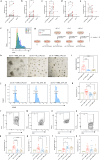
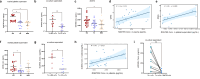
Myasthenia gravis (MG) is an autoimmune disorder that disrupts neuromuscular junction function through autoantibodies. Platelets are emerging as key players in the pathogenesis of MG, bridging innate and adaptive immunity. We analyze platelet transcriptome signatures and their interactions with the immune system in AChR+ immunotherapy-naïve MG (nMG) patients using bulk and single-cell RNA sequencing on peripheral blood mononuclear cells (PBMC). nMG patients exhibit upregulation of genes related to activation, inflammation, and cytoskeletal regulation. Increased platelet count, activation, altered morphology, enhanced CD62P expression, and elevated plasma CD40L levels are observed in PBMCs, which diminish with minimal clinical status (MMS). Functionally, platelets show heightened interactions with leukocytes, forming aggregates that correlate with disease severity. These features return to baseline after intravenous immunoglobulin or prolonged immunosuppressive therapy. This study underscores platelet activation's critical role in MG and supports platelet-targeted therapy.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- Huijbers, M. G., Marx, A., Plomp, J. J., Le Panse, R. & Phillips, W. D. Advances in the understanding of disease mechanisms of autoimmune neuromuscular junction disorders. Lancet Neurol.21, 163–175 (2022). - PubMed
-
- Villegas, J. A., Van Wassenhove, J., Le Panse, R., Berrih-Aknin, S. & Dragin, N. An imbalance between regulatory T cells and T helper 17 cells in acetylcholine receptor-positive myasthenia gravis patients. Ann. N. Y. Acad. Sci.1413, 154–162 (2018). - PubMed
-
- Zhao, R., Luo, S. & Zhao, C. The role of innate immunity in myasthenia gravis. Autoimmun. Rev.20, 102800 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

