Trans-eQTL mapping prioritises USP18 as a negative regulator of interferon response at a lupus risk locus
- PMID: 41038828
- PMCID: PMC12491431
- DOI: 10.1038/s41467-025-63856-7
Trans-eQTL mapping prioritises USP18 as a negative regulator of interferon response at a lupus risk locus
Abstract
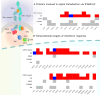
Although genome-wide association studies have provided valuable insights into the genetic basis of complex traits and diseases, translating these findings to causal genes and their downstream mechanisms remains challenging. We performed trans expression quantitative trait locus (trans-eQTL) meta-analysis in 3734 lymphoblastoid cell line samples, identifying four robust loci that replicated in an independent multi-ethnic dataset of 682 individuals. The trans-eQTL signal at the ubiquitin specific peptidase 18 (USP18) locus colocalised with a GWAS signal for systemic lupus erythematosus (SLE). USP18 is a known negative regulator of interferon signalling and the SLE risk allele increased the expression of 50 interferon-inducible genes, suggesting that the risk allele impairs USP18's ability to effectively limit the interferon response. Intriguingly, the USP18 trans-eQTL signal would not have been discovered in a meta-analysis of up to 43,301 whole blood samples, reaffirming the importance of capturing context-specific genetic effects for GWAS interpretation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.C. is an employee of Pfizer. E.R.H., M.C.T., and J.C.M. are employees of Bristol Myers Squibb. H.O. is an employee of GSK. N.N. was an employee of GSK while this work was conducted. The other authors declare no competing interests.
Figures


References
-
- Aguet, F. et al. Molecular quantitative trait loci. Nat. Rev. Methods Prim.3, 1–22 (2023).
-
- Frangoul, H. et al. CRISPR-Cas9 gene editing for sickle cell disease and β-Thalassemia. N. Engl. J. Med.384, 252–260 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- PRG1117/Eesti Teadusagentuur (Estonian Research Council)
- 310030_152724/1/Swiss National Science Foundation | National Center of Competence in Research Affective Sciences - Emotions in Individual Behaviour and Social Processes (National Centre of Competence in Research Affective Sciences)
- PSG415/Eesti Teadusagentuur (Estonian Research Council)
- MOB3ERC115/Eesti Teadusagentuur (Estonian Research Council)
- 220540/Z/20/A/Wellcome Trust (Wellcome)
LinkOut - more resources
Full Text Sources
Medical

