RSL3 promotes PARP1 apoptotic functions by distinct mechanisms during ferroptosis
- PMID: 41039223
- PMCID: PMC12492750
- DOI: 10.1186/s11658-025-00785-9
RSL3 promotes PARP1 apoptotic functions by distinct mechanisms during ferroptosis
Abstract
Background: The classical ferroptosis activator RSL3 targets enzymes with nucleophilic active sites, primarily acting on glutathione peroxidase 4 (GPX4) to trigger ferroptosis. Recent studies identify RSL3 as a potential pro-apoptotic agent. However, the mechanism by which RSL3 induces apoptosis during ferroptosis remains elusive. Poly(ADP-ribose) polymerase (PARP1) determines cell fate in response to DNA damage, where its loss or cleavage by activated caspase-3 induces apoptosis to attenuate tumor progression. We elucidate a novel mechanism underlying PARP1 regulation, encompassing both its caspase-dependent cleavage and full-length depletion during RSL3-mediated ferroptosis-apoptosis crosstalk.
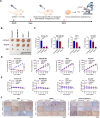
Methods: To investigate the role of RSL3 during ferroptosis, we treated several cancer cells of different histological types with varying doses of RSL3 to induce apoptosis. The regulatory proteins of PARP1 were analyzed using real-time quantitative polymerase chain reaction (RT-qPCR) and Western blot analysis. The N6-methyladenosine (m6A) modification level of PARP1 was determined by m6A RNA immunoprecipitation (MeRIP)-qPCR analysis. Additionally, an RNA immunoprecipitation (RIP)-qPCR assay was performed to identify the target protein of the m6A site of PARP1. Furthermore, we established a mouse xenograft model of PARP inhibitor (PARPi)-resistant cells to analyze the effect of RSL3 on PARPi-resistant tumor growth.
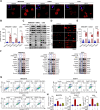
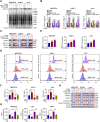
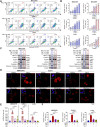
Results: RSL3 triggers two parallel apoptotic pathways via increasing reactive oxygen species (ROS) production during ferroptosis: (1) caspase-dependent PARP1 cleavage and (2) DNA damage-dependent apoptosis resulting from reduced full-length PARP1. The latter occurs through inhibition of METTL3-mediated m6A modification and subsequent suppression of PARP1 translation. Moreover, we found that RSL3 retains pro-apoptotic functions in PARPi-resistant cells and effectively inhibits PARPi-resistant xenograft tumor growth in vivo.
Conclusions: RSL3 orchestrates ferroptosis-apoptosis crosstalk via PARP1, demonstrating therapeutic potential against tumorigenesis, particularly in PARPi-resistant malignancies.
Keywords: N 6-methyladenosine; Apoptosis; PARP1; PARPi-resistant tumor; RSL3.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All animal protocols in this study were performed in accordance with the Institutional Animal Care and Use Committee, University Laboratory Animal Research of Wenzhou Medical University (no. wydw2024-0522, 25 January 2024) that acts on the International Council for Laboratory Animal Science (ICLAS). Consent for publication: All the authors have provided their consent for publication of the manuscript. Competing interests: The authors have no conflicts of interest to declare.
Figures



References
-
- Landes T, Martinou JC. Mitochondrial outer membrane permeabilization during apoptosis: the role of mitochondrial fission. Biochim Biophys Acta. 2011;1813(4):540–5. - PubMed
-
- Costa I, Barbosa D, Benfeito S, Silva V, Chavarria D, Borges F, et al. Molecular mechanisms of ferroptosis and their involvement in brain diseases. Pharmacol Ther. 2023;244: 108373. - PubMed
-
- Huang C, Yang M, Deng J, Li P, Su W, Jiang R. Upregulation and activation of p53 by erastin-induced reactive oxygen species contribute to cytotoxic and cytostatic effects in A549 lung cancer cells. Oncol Rep. 2018;40(4):2363–70. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

