A comparative proteomic, transcriptomic and glycomic analysis of extracellular vesicle isolation techniques highlights ExoGAG efficiency for a more complete identification of breast milk molecular signaling pathways
- PMID: 41039499
- PMCID: PMC12492545
- DOI: 10.1186/s12964-025-02390-x
A comparative proteomic, transcriptomic and glycomic analysis of extracellular vesicle isolation techniques highlights ExoGAG efficiency for a more complete identification of breast milk molecular signaling pathways
Abstract
Background: Human milk (HM) is the first form of communication between mothers and newborns and it is implicated in the infant growth and protection. We recently showed a functional characterization of HM, unmasking the molecular mechanisms related to EVs signaling and its functional role in prematurity. In that study, we identified the need to establish and optimize a standard isolation protocol for human milk extracellular vesicles (mEVs).
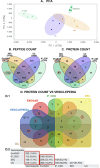
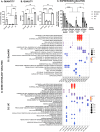
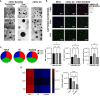
Methods: Four mEVs isolation methods were compared: ultracentrifugation (UC), size exclusion chromatography (SEC), immunoprecipitation with tetraspanin CD9 (IP_CD9) and ExoGAG. Three pools of human milk (each composed of samples from ten donor mothers) were used and isolation of mEVs was performed starting from the same volume for each method. The proteomic, transcriptomic and glycomic composition of the extracellular vesicles obtained after isolation was then analyzed for each method. The sensitivity, specificity and quality of the results were also determined. A comparative analysis of results common across all isolation methods was performed to identify potential signaling pathways associated with mEVs.
Results: ExoGAG and UC proved to be the most efficient of the four techniques compared for mEVs isolation. However, ExoGAG compared to UC provided a higher concentration of total and vesicle-related proteins and peptides and a higher glycoprotein count keeping all the glycan subgroups. Despite ExoGAG and UC show similar vesicle profiles in terms of size, concentration, tetraspanin subpopulations and EVs markers, ExoGAG was the most efficient technique in terms of accuracy, consistency and reproducibility for omics studies. Furthermore, results allowed us to identify that mEVs components are involved in the signaling pathways of infant biological development, immune system maturation and protein metabolism.
Conclusions: This study establishes UC and ExoGAG as reliable methods for mEVs isolation and describes its protocol, being ExoGAG the most efficient. Also, the omics analysis show that biomolecules conforming those mEVs are linked to the defense system against external agents (specific role in the immune system pathway) and in the correct establishment of the neural structure (developmental pathway), while providing all the nutritional requirements for the correct growth of the newborn (metabolic pathway).
Graphical Abstract:
Supplementary Information: The online version contains supplementary material available at 10.1186/s12964-025-02390-x.
Keywords: Extracellular vesicles; Glycomics; Human milk; Isolation method; Proteomics; Transcriptomics.
Conflict of interest statement
Declarations. Ethics approval and informed consent to participate: The participating mothers gave their written informed consent to participate in this study. Ethics committee approval was obtained from Research Ethics Committees of Galicia (register code: 2020/243). Competing interests: Only Miguel A. Garcia Gonzalez reports conflict of interest as is included as author in the patent of the isolation method KITGAG (Commercialized as ExoGAG).
Figures







References
-
- Basak T, Bhat A, Malakar D, Pillai M, Sengupta S. In-depth comparative proteomic analysis of yeast proteome using iTRAQ and SWATH based MS. Mol Biosyst. 2015;11(8):2135–43. 10.1039/c5mb00234f. - PubMed
Grants and funding
- RD21/0005/0020/Instituto de Salud Carlos III (ISCIII) RICORS program to RICORS2040
- RD21/0005/0020/Instituto de Salud Carlos III (ISCIII) RICORS program to RICORS2040
- RD21/0005/0020/Instituto de Salud Carlos III (ISCIII) RICORS program to RICORS2040
- RD21/0005/0020/Instituto de Salud Carlos III (ISCIII) RICORS program to RICORS2040
- PI22/00227/Instituto de Salud Carlos III (ISCIII)
- PI22/00227/Instituto de Salud Carlos III (ISCIII)
- PI15/01467/Instituto de Salud Carlos III (ISCIII)
- PI15/01467/Instituto de Salud Carlos III (ISCIII)
- PI15/01467/Instituto de Salud Carlos III (ISCIII)
- SEN2021_2/Senefro foundation
- SEN2021_2/Senefro foundation
- SEN2021_2/Senefro foundation
- SEN2021_2/Senefro foundation
- SEN2021_2/Senefro foundation
- IN607B2023/07/Xunta de Galicia/GAIN , Grupos con potencial de crecemento
- IN607B2023/07/Xunta de Galicia/GAIN , Grupos con potencial de crecemento
- IN607B2023/07/Xunta de Galicia/GAIN , Grupos con potencial de crecemento
- IN607B2023/07/Xunta de Galicia/GAIN , Grupos con potencial de crecemento
- IN607B2023/07/Xunta de Galicia/GAIN , Grupos con potencial de crecemento
LinkOut - more resources
Full Text Sources

