WUSCHEL-dependent chromatin regulation in maize inflorescence development at single-cell resolution
- PMID: 41039501
- PMCID: PMC12492591
- DOI: 10.1186/s13059-025-03791-4
WUSCHEL-dependent chromatin regulation in maize inflorescence development at single-cell resolution
Abstract
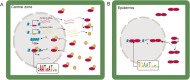
Background: WUSCHEL (WUS) is a homeodomain transcription factor vital for stem cell proliferation in plant meristems. In maize, ZmWUS1 is expressed in the inflorescence meristem, including the central zone reservoir of stem cells. ZmWUS1 overexpression in the Barren inflorescence3 (Bif3) mutant perturbs inflorescence development due to stem cell over-proliferation.
Results: Single-cell Assay for Transposase Accessible Chromatin sequencing (scATAC-seq) shows that Bif3 alters central zone chromatin accessibility compared to normal inflorescence meristems. The CAATAATGC motif, a known homeodomain recognition site, is enriched within regions with increased chromatin accessibility in Bif3, suggesting ZmWUS1 could function as a transcriptional activator in the central zone. This motif differs from the TGAATGAA motif identified by DNA Affinity Purification sequencing (DAP-seq) of ZmWUS1, which showed low enrichment in the central zone. Conversely, regions with decreased chromatin accessibility in Bif3 are instead adjacent to AUXIN RESPONSE FACTOR genes, suggesting possible reduced auxin signaling in the Bif3 central zone.
Conclusions: This study characterized how Bif3 overexpression of ZmWUS1 influences chromatin accessibility in the central zone, reducing auxin signaling, while raising questions about differential ZmWUS1 motif usage in distinct cellular contexts.
Keywords: Cis-regulatory elements; WUSCHEL; ZmWUS1; Epigenomics; Inflorescence meristem; Maize ear; Meristem development; Single-cell ATAC-seq.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Wenjing She was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team. The peer-review history is available in the online version of this article. Consent for publication: Not applicable. Competing interests: R.J.S. is a co-founder of REquest Genomics, LLC, a company that provides epigenomic services. The remaining authors declare no competing interests.
Figures





References
-
- Laux T, Mayer KF, Berger J, Jürgens G. The wuschel gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. - PubMed
-
- Clark SE, Running MP, Meyerowitz EM. CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development. 1995;121:2057–67.
-
- Schoof H, Lenhard M, Haecker A, Mayer KF, Jürgens G, Laux T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–44. - PubMed
-
- Suzaki T, Toriba T, Fujimoto M, Tsutsumi N, Kitano H, Hirano H-Y. Conservation and diversification of meristem maintenance mechanism in Oryza sativa: function of the FLORAL ORGAN NUMBER2 gene. Plant Cell Physiol. 2006;47:1591–602. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

