BRD9-p53-E2F1 circuit orchestrates cell growth and DNA damage repair in gastric cancer
- PMID: 41039535
- PMCID: PMC12492862
- DOI: 10.1186/s12943-025-02449-w
BRD9-p53-E2F1 circuit orchestrates cell growth and DNA damage repair in gastric cancer
Abstract
Background: BRD9 is involved in multiple physiological and pathological pathways, yet its functional role and molecular mechanisms in gastric cancer (GC) remain largely unexplored. Addressing this knowledge gap is critical given the persistent global mortality burden of GC and the limited efficacy of current therapeutic strategies.
Methods: BRD9 expression in GC patients was systematically analyzed using immunohistochemical (IHC) assays and transcriptomic datasets. Comprehensive functional validation, employing cellular and murine tumor models, elucidated BRD9's role in GC progression. Molecular pathways underlying BRD9-mediated gastric carcinogenesis were delineated through integrated approaches, including RNA sequencing, co-immunoprecipitation (co-IP), subcellular fractionation, and luciferase reporter assays.
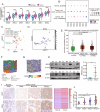
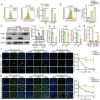
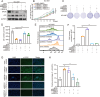
Results: BRD9 was significantly overexpressed in GC and associated with poor patient prognosis. Functionally, BRD9 promoted GC cell proliferation and enhanced DNA damage repair capacity. Mechanistically, elevated BRD9 expression inhibited p53 nuclear translocation via direct binding, subsequently activating the E2F transcription factor family. Notably, we identified that E2F1 directly bound to and transactivated the BRD9 promoter, establishing a positive feedback loop that sustains BRD9 expression. Additionally, BRD9 knockdown sensitized GC cells to cisplatin and oxaliplatin treatment.
Conclusions: These findings highlight the critical role of BRD9 in GC progression and its therapeutic potential. The BRD9-p53-E2F1 axis acts as a crucial regulator of GC cell proliferation and DNA damage response. Targeting BRD9 pharmacologically could be a novel therapeutic approach to enhance chemotherapy efficacy and improve treatment outcomes in GC patients.
Keywords: BRD9; DNA damage repair; E2F family; Gastric cancer; P53; Progression.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The clinical samples and information used in the study was approved by the Institutional Review Board of Fudan University Shanghai Cancer Center. The animal experiments were conducted in compliance with protocols approved by the Shanghai Medical Experimental Animal Care Commission. Competing interests: The authors declare no competing interests.
Figures






References
-
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74:12–49. - PubMed
-
- Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–48. - PubMed
-
- López MJ, Carbajal J, Alfaro AL, Saravia LG, Zanabria D, Araujo JM, et al. Characteristics of gastric cancer around the world. Crit Rev Oncol Hematol. 2023;181:103841. - PubMed
-
- Alsina M, Arrazubi V, Diez M, Tabernero J. Current developments in gastric cancer: from molecular profiling to treatment strategy. Nat Rev Gastroenterol Hepatol. 2023;20:155–70. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

