Identification of a PRDM1-regulated T cell network to regulate atherosclerotic plaque inflammation
- PMID: 41039608
- PMCID: PMC12490039
- DOI: 10.1186/s13073-025-01541-6
Identification of a PRDM1-regulated T cell network to regulate atherosclerotic plaque inflammation
Abstract
Background: Inflammation is a key driver of atherosclerosis, yet the mechanisms sustaining inflammation in human plaques remain poorly understood. This study uses a network-based approach to identify immune gene programs involved in the transition from low- to high-risk (rupture-prone) human atherosclerotic plaques.
Methods: Expression data from human carotid artery plaques, both stable (low-risk, n = 16) and unstable (high-risk, n = 27), were analyzed using Weighted Gene Co-expression Network Analysis (WGCNA). Bayesian network inference, operated on the eigengene values from the WGCNA, further extended the WGCNA analysis, and similarity to the signature of T cell subsets was validated in single-cell RNA sequencing data of human plaques, and a loss-of-function study in a mouse model of atherosclerosis. In silico drug repurposing was performed to identify potential therapeutic targets.
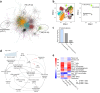
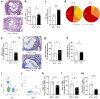
Results: Our analysis revealed a distinct gene module with a prominent T cell signature, particularly in unstable plaques. Key regulatory factors, RUNX3, IRF7 and in particular PRDM1, were significantly downregulated in plaque T cells from symptomatic versus asymptomatic patients, indicating a protective role. Additionally, as PRDM1 is downstream of IRF7, we opted for PRDM1 as a key target. T cell-specific Prdm1 deficiency in Western-type diet fed Ldlr knockout mice featured accelerated plaque progression. Finally, as PRDM1 targeting drugs are not yet available, we performed in silico drug repurposing, identifying EGFR inhibitors as promising therapeutic candidates.
Conclusions: This study highlights a PRDM1-regulated T cell network that distinguishes high-risk from low-risk plaques and demonstrates the regulatory role of T cell PRDM1 in controlling atherosclerosis, positioning this pathway as a promising therapeutic target.
Keywords: Atherosclerosis; Carotid endarterectomy; Microarray; PRDM1; Single-cell sequencing; T cell.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All patient materials collected for the MaasHPS dataset (GSE163154) [18] were obtained in accordance with the Dutch Code for Proper Secondary Use of Human Tissue ( https://www.federa.org ) and with approval from the local Medical Ethical Committee (protocol number 16-4-181). This study conforms to the principles of the Declaration of Helsinki, and all participants provided written informed consent prior to inclusion. All BiKE human samples (GSE21545) [21] were collected with informed consent from patients or guardians of organ donors. BiKE studies were approved by the Karolinska Institute ethics committee (file numbers 02-147 and 2009/295-31/2) and were conducted in accordance with the Declaration of Helsinki. Due to GDPR and ethical regulations protecting participant privacy, individual-level human data cannot be deposited or shared. Three patients with severe carotid plaque requiring carotid endarterectomy were identified and consented in collaboration with the Scripps Health Biorepository, under IRB# 19-7332 approved by the Scripps Institutional Review Board. Informed consent was obtained for all collected samples, which were used for the scRNA-seq dataset (GSE159677) [23]. Patients undergoing carotid endarterectomy at Mount Sinai Hospital were enrolled in an ongoing clinical study approved by the Institutional Review Board of the Icahn School of Medicine at Mount Sinai, under IRB# 11–01427. Written informed consent was obtained from all eligible participants. Exclusion criteria included current infection, autoimmune disease, active or recurrent cancer, severe renal failure requiring dialysis, or peripheral arterial occlusive disease causing pain at rest. Samples were collected from each patient and used to generate the scRNA-seq dataset (GSE224273) [25]. All mouse experiments were approved by the regulatory authority of the Maastricht University Medical Centre (permit number: 2018-011-008) and local German regulatory authorities (Landesamt für Natur, Umwelt und Verbraucherschutz Nordrhein-Westfalen, Germany, approval number 81-02.04.2019.A363). The experiments complied with the Dutch governmental guidelines and Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes and the German animal protection law. Furthermore, the study was conducted in compliance with the ARRIVE guidelines. Every effort was made to minimize suffering. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




References
-
- Libby P, Ridker PM, Hansson GK. Progress and challenges in translating the biology of atherosclerosis. Nature. 2011;473(7347):317–25. - PubMed
-
- Shah PK. Mechanisms of plaque vulnerability and rupture. J Am Coll Cardiol. 2003;41(4, Supplement):S15-22.
-
- Bentzon JF, Otsuka F, Virmani R, Falk E. Mechanisms of plaque formation and rupture. Circ Res. 2014;114(12):1852–66. - PubMed
-
- Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med. 2017;377(12):1119–31. - PubMed
MeSH terms
Substances
Grants and funding
- 2023KJ107/Tianjin Municipal Education Commission Scientific Research Plan Project
- 201609120004/China Scholarship Council
- 202308500067/China Scholarship Council
- 201706990018/China Scholarship Council
- 202106100042/China Scholarship Council
- 2019-02027, 2023-02724/Vetenskapsrådet
- 20241083, 20240094, 20210466, 20200621, 20200520, 20180244, 20180247, 201602877/Hjärt-Lungfonden
- P13-0171/Svenska Sällskapet för Medicinsk Forskning
- PG/21/10541, PG/21/10634, PG/21/10557 and PG/24/11946/BHF_/British Heart Foundation/United Kingdom
- 2022T45AXH/Ministero dell'Istruzione, dell'Università e della Ricerca
- 2016T060/Hartstichting
- 15CVD04/Fondation Leducq
- 10.20.2.043MN/Fritz Thyssen Stiftung
- 91619053/Nederlandse Organisatie voor Wetenschappelijk Onderzoek
- 0.16.186.364/Nederlandse Organisatie voor Wetenschappelijk Onderzoek
- JTC-2017t100 AtheroMacHete/European Research Area Network on Cardiovascular Diseases
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

