This is a preprint.
Mechanisms of superior respiratory IgA responses against SARS-CoV-2 after mucosal vaccination
- PMID: 41040255
- PMCID: PMC12485830
- DOI: 10.1101/2025.09.22.677712
Mechanisms of superior respiratory IgA responses against SARS-CoV-2 after mucosal vaccination
Abstract
Mucosal immunization and respiratory IgA offer significant promise in protecting against airborne pathogens, including SARS-CoV-2. However, the conditions and mechanisms that lead to the robust induction of respiratory IgA responses following mucosal vaccination remain poorly understood. It is also currently debatable whether mucosal vaccination is still warranted given that most individuals in developed countries have established a hybrid immunity from vaccination and infection. Here we characterized respiratory mucosal immune responses after SARS-CoV-2 infection, vaccination or both in humans. We found that hybrid immunity resulted in moderately increased respiratory IgA and neutralizing antibody responses compared to infection or vaccination alone. However, a direct comparison of hybrid immunity and a mucosal adenovirus-based booster vaccination in animal models revealed that respiratory booster immunization elicited markedly stronger and more durable respiratory IgA, T cell response, and protective immunity against SARS-CoV-2, supporting the promise of respiratory mucosal vaccination. Mechanistically, we found that mucosal booster immunization induced local IgA-secreting cells in the respiratory mucosa, aided by pulmonary CD4+ T cells in situ. Strikingly, local IL-21-producing Blimp-1+ Th1 effector cells were critical in mediating the CD4+ T cell help for respiratory IgA production. Furthermore, lung macrophages were important for this respiratory IgA response via the production of TGF-β. Consequently, we demonstrated delivery of adenoviral booster to the lower airway was necessary to generate robust upper and lower airway IgA responses. Collectively, our results uncover a local cellular network supporting enhanced respiratory IgA responses, with implications for the development of optimal mucosal immunization strategies against SARS-CoV-2 and other respiratory pathogens.
Figures






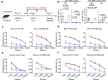



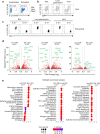
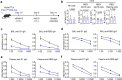










References
Publication types
Grants and funding
- S10 OD028518/OD/NIH HHS/United States
- U54 CA260582/CA/NCI NIH HHS/United States
- F31 HL170746/HL/NHLBI NIH HHS/United States
- T32 AI007496/AI/NIAID NIH HHS/United States
- R01 AI176171/AI/NIAID NIH HHS/United States
- R01 AI147394/AI/NIAID NIH HHS/United States
- R01 HL170961/HL/NHLBI NIH HHS/United States
- R01 AI112844/AI/NIAID NIH HHS/United States
- R01 AI154598/AI/NIAID NIH HHS/United States
- T32 CA009109/CA/NCI NIH HHS/United States
- R01 AI168684/AI/NIAID NIH HHS/United States
- R01 AG069264/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
