This is a preprint.
Targeting RAD52 overcomes PARP inhibitor resistance in preclinical Brca2-deficient ovarian cancer model
- PMID: 41040355
- PMCID: PMC12485677
- DOI: 10.1101/2025.09.24.678351
Targeting RAD52 overcomes PARP inhibitor resistance in preclinical Brca2-deficient ovarian cancer model
Abstract
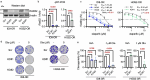
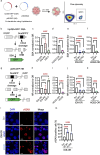
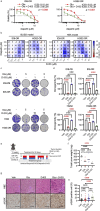
BRCA-mutated ovarian cancer commonly develops resistance to poly (ADP-ribose) polymerase (PARP) inhibitors. Here, we investigated the DNA repair protein RAD52 as a potential target to overcome resistance. In analysis of The Cancer Genome Atlas datasets and immunohistochemistry of tissue microarrays, elevated RAD52 expression correlated with poor overall survival in patients with high-grade serous ovarian cancers. We tested two PARP inhibitor-resistant Brca2-deficient mouse ovarian cancer models, ID8-OR and HGS2-OR. HGS2-OR cells had higher RAD52 expression than parental lines. Rad52 knockout or knockdown restored PARP inhibitor sensitivity in both models. In syngeneic mice, ID8-OR cells in which Rad52 was knocked out yielded lower tumor burden and longer overall survival than control cells. Rad52 depletion impaired single-strand annealing and homologous recombination and led to accumulation of DNA double-strand breaks after PARP inhibitor treatment. RNA sequencing demonstrated that PARP inhibitor treatment induced Polq expression in Brca2- and Rad52-deficient cells, suggesting a switch to microhomology-mediated end joining. Finally, the RAD52 inhibitor D-I03 synergized with a PARP inhibitor to reduce cell viability and tumor burden and prolong survival. Collectively, our findings establish RAD52 as a promising therapeutic target to overcome PARP inhibitor resistance in BRCA2-mutated ovarian cancer and offer mechanistic insights to inform future clinical strategies.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures



References
-
- Gilks C. B. Subclassification of ovarian surface epithelial tumors based on correlation of histologic and molecular pathologic data. Int J Gynecol Pathol 23, 200–205 (2004). - PubMed
-
- Seidman JD, H.-S I., Haiba M, Boice CR, Kurman RJ, Ronnett BM. The histologic type and stage distribution of ovarian carcinomas of surface epithelial origin. Int J Gynecol Pathol 23, 41–44 (2004). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
