Refined and benchmarked homemade media for cost-effective, weekend-free human pluripotent stem cell culture
- PMID: 41040810
- PMCID: PMC12488189
- DOI: 10.12688/openreseurope.18245.2
Refined and benchmarked homemade media for cost-effective, weekend-free human pluripotent stem cell culture
Abstract
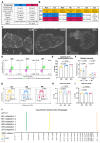
Background: Cost-effective, practical, and reproducible culture of human pluripotent stem cells (hPSCs) is required for basic and translational research. Basal 8 (B8) has emerged as a cost-effective solution for weekend-free and chemically-defined hPSC culture. However, the requirement to home-produce some recombinant growth factors for B8 can hinder access and reproducibility. Moreover, we found the published B8 formulation suboptimal in widely-used normoxic hPSC culture. Lastly, the performance of B8 in functional applications such as genome editing or organoid differentiation required systematic evaluation.
Methods: We formulated B8 with commercially available, growth factors and adjusted its composition to support normoxic culture of WTC11 human induced pluripotent stem cell line. We compared this formulation (B8+) with commercial Essential 8 (cE8) and a home-made, weekend-free E8 formulation (hE8). We measured pluripotency marker expression and cell cycle by flow cytometry, and investigated the transcriptional profiles by bulk and single-cell RNA sequencing. We further assessed genomic stability, genome editing efficiency, single-cell cloning, and differentiation in both monolayer and organoids. Finally, we validated key findings using male (H1) and female (H9) human embryonic stem cells.
Results: hE8 performed comparably to cE8 across most functional assays and cell lines. In contrast, cells in B8+ displayed higher NANOG expression and improved genome editing efficiency. At the same time, B8+ led to gene expression changes indicative of marked lineage priming, reflected in altered morphology and differential response to some differentiation protocols. Both weekend-free media resulted in a modest transcriptional shift towards a less metabolically active state, consistent with intermittent media starvation.
Conclusions: Homemade weekend-free media can provide a cost-effective alternative to commercial formulations. hE8, integrating some features of B8 while resembling cE8, emerges as a robust and practical option with limited compromises. B8+, though advantageous in some contexts, warrants caution due to lineage priming effects that may impact differentiation outcomes.
Keywords: hiPSC; pluripotency; culture media; thermostable FGF2; TGF beta.
Copyright: © 2025 Truszkowski L et al.
Conflict of interest statement
Competing interests: H.B. and L.F. are employees of Qkine and C.E. was the CEO of Qkine for most of the study. The rest of the authors declare no competing interests.
Figures






References
LinkOut - more resources
Full Text Sources
Research Materials
