Hsa_circ_0058495-mediated IGF2BP2 ubiquitination and m6A modification of MEKK1 promote the progression of PDAC
- PMID: 41041073
- PMCID: PMC12486403
- DOI: 10.7150/thno.117202
Hsa_circ_0058495-mediated IGF2BP2 ubiquitination and m6A modification of MEKK1 promote the progression of PDAC
Abstract
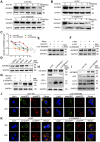
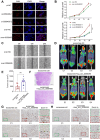
Background: Pancreatic ductal adenocarcinoma (PDAC) is a highly aggressive malignancy with dismal clinical outcomes. We identified hsa_circ_0058495 as significantly upregulated in PDAC tissues and PDAC cell-derived exosomes, where it contributes to tumor proliferation and invasion. The molecular mechanisms underlying its oncogenic function, however, remain incompletely understood. Methods: Differential circRNA expression was profiled by RNA sequencing. The functional role of hsa_circ_0058495 and its molecular interactions were interrogated through Western blotting, RT-qPCR, co-immunoprecipitation, RNA pull-down, and RNA immunoprecipitation assays. Confocal microscopy and PET/CT imaging were employed to delineate its biological effects in vitro and in vivo. Results: Hsa_circ_0058495 was enriched in PDAC-derived exosomes and stabilized IGF2BP2 by preventing TRIM25-mediated ubiquitination and attenuating autophagy-dependent degradation. Stabilized IGF2BP2 enhanced the stability of MEKK1 mRNA, leading to sustained ERK1/2 phosphorylation and consequent promotion of PDAC cell proliferation and invasion. Moreover, exosomal hsa_circ_0058495 facilitated M2 macrophage polarization, thereby fostering an immunosuppressive tumor microenvironment. Conclusions: Hsa_circ_0058495 promotes PDAC progression by stabilizing IGF2BP2 and activating the MEKK1-ERK signaling cascade, while exosomal transfer of hsa_circ_0058495 drives M2 macrophage polarization to reinforce tumor-associated immunosuppression. These findings establish hsa_circ_0058495 as a pivotal regulator of PDAC progression and underscore its potential utility as both a diagnostic biomarker and a therapeutic target.
Keywords: M2 polarization; PDAC; circRNA; m6A modification; ubiquitination.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures





References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74:229–63. - PubMed
-
- Springfeld C, Ferrone CR, Katz MHG, Philip PA, Hong TS, Hackert T. et al. Neoadjuvant therapy for pancreatic cancer. Nat Rev Clin Oncol. 2023;20:318–37. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

