This is a preprint.
RPS19 and RPL5 Haploinsufficient Models Reveal Divergent Ribosomal Subunit Controls of Fetal Hematopoiesis
- PMID: 41041537
- PMCID: PMC12486077
- DOI: 10.21203/rs.3.rs-7563799/v1
RPS19 and RPL5 Haploinsufficient Models Reveal Divergent Ribosomal Subunit Controls of Fetal Hematopoiesis
Abstract
Diamond Blackfan anemia syndrome (DBAS) is a congenital ribosomopathy caused by haploinsufficiency of ribosomal proteins (RPs), but how RP stoichiometry and activity regulates erythroid development remains enigmatic. Using novel in vivo models, we uncover strikingly divergent functions for the small and large ribosomal subunit proteins RPS19 and RPL5 in fetal hematopoiesis. While RPL5 haploinsufficiency causes hematopoietic stem and progenitor cell (HSPC) accumulation and prenatal lethality via p53-mediated ferroptosis of mature erythroid progenitors, RPS19 haploinsufficiency leads to HSPC depletion and impaired erythroid expansion through p53-dependent apoptosis. The latter is accompanied by translational and transcriptional dysregulation, including the upregulation of RUNX1, which is also observed in RPS- haploinsufficient DBAS patients. Importantly, Runx1 deletion in RPS19-haploinsufficient mice partially rescues HSPC numbers. These findings reveal subunit-specific RP functions in controlling fetal hematopoiesis and demonstrate how imbalanced RP stoichiometry disrupts developmental programs, providing crucial mechanistic insights into DBAS pathogenesis and the basis for its clinical heterogeneity.
Conflict of interest statement
DECLARATION OF INTERESTS The authors declare no competing interests.
Figures


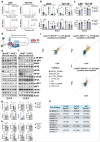
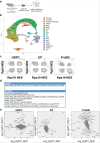



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

