Limited phosphate resources are efficiently allocated to each organ by the PHR1-EIN3/EIL1 module to coordinate their establishment
- PMID: 41042860
- PMCID: PMC12494022
- DOI: 10.1126/sciadv.adw7727
Limited phosphate resources are efficiently allocated to each organ by the PHR1-EIN3/EIL1 module to coordinate their establishment
Abstract
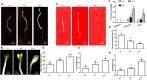
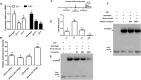
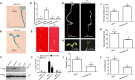
Phosphate (Pi) is an essential nutrient that frequently limits plant growth because of its low availability in soils, especially during early seedling development in Arabidopsis. Under Pi-deficient conditions, etiolated seedlings exhibit elongated hypocotyls, shortened roots, and small, pale cotyledons-a morphological adaptation that enhances light-foraging capacity. However, how Pi is strategically distributed among these organs to optimize seedling establishment remains unclear. We here identify a PHR1-EIN3/EIL1 regulatory module that directs minimal Pi resources to suppress root elongation and cotyledon expansion or to accelerate greening, promoting Pi conservation. This mechanism prioritizes hypocotyl elongation to improve light acquisition. In contrast, under sufficient Pi supply, hypocotyl and root growth is promoted along with cotyledon enlargement, while greening is delayed. Thus, the PHR1-EIN3/EIL1 module enables efficient allocation of limited Pi to maximize hypocotyl elongation for light foraging during early seedling establishment while strategically restricting Pi usage in other organs to enhance overall survival.
Figures




References
-
- Mu X. R., Tong C., Fang X. T., Bao Q. X., Xue L. N., Meng W. Y., Liu C. Y., Loake G. J., Cao X. Y., Jiang J. H., Meng L. S., Feed-back loop promotes sucrose accumulation in cotyledons to facilitate sugar-ethylene signaling-mediated, etiolated-seedling greening. Cell Rep. 38, 110529 (2022). - PubMed
-
- Mu X. R., Wang Y. B., Bao Q. X., Wei Y. T., Zhao S. T., Tao W. Z., Liu Y. X., Wang W. N., Yu F. H., Tong C., Wang J. W., Gu C. Y., Wang Q. M., Liu X. R., Sai N., Zhu J. L., Zhang J., Loake G. J., Meng L. S., Glucose status within dark-grown etiolated cotyledons determines seedling de-etiolation upon light irradiation. Plant Physiol. 194, 391–407 (2024). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

