Distinct T cell functions enable efficient immunoediting and prevent tumor emergence of developing sarcomas
- PMID: 41043437
- PMCID: PMC12693725
- DOI: 10.1016/j.ccell.2025.09.005
Distinct T cell functions enable efficient immunoediting and prevent tumor emergence of developing sarcomas
Abstract
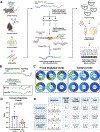
T cells edit tumors by eliminating neoantigen-expressing tumor cells. Yet, how and when this is achieved remains uncertain. Using a murine sarcoma model with fluorescent neoantigens, we found that tumors developed later and in fewer T cell-sufficient mice (∼53% penetrance) than T cell-deficient mice (∼100%). With T cells, all emergent tumor cells had silenced neoantigens, but neoantigen-negative tumor cells were also present in every T cell-deficient mouse. This suggested silencing was necessary but not sufficient for outgrowth. Genetic removal of neoantigens restored tumor penetrance if implemented on day 5 post-tumor initiation, but not day 10, because CD8+ and CD4+ T cells infiltrated the tissue and eliminated most neoantigen-positive and -negative tumor cells within 8 days. Single-cell analyses on day-7 tumors showed oncogenic changes including increased proliferation and T cell-dependent upregulation of the IFNγ-response gene Cd274 (PD-L1). T cell-depletion rescued both neoantigen-positive and -negative cells, while IFNγ blockade rescued only negative cells. This shows that T cells efficiently edit sarcomas of neoantigens and prevent early tumors via IFNγ-independent and IFNγ-dependent (bystander) mechanisms.
Keywords: GEMMs; Immunoediting; T cell elimination; escape; immunogenicity; immunosurveillance; neoantigen silencing; pre-emergent tumor; sarcoma; tumor clonality.
Copyright © 2025 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
