Muscle-derived miR-126 regulates TDP-43 axonal local synthesis and NMJ integrity in ALS models
- PMID: 41044342
- PMCID: PMC12586178
- DOI: 10.1038/s41593-025-02062-6
Muscle-derived miR-126 regulates TDP-43 axonal local synthesis and NMJ integrity in ALS models
Abstract
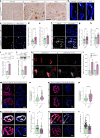
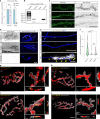
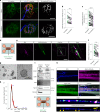
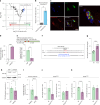
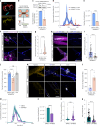
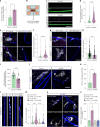
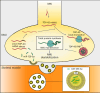
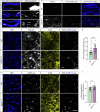
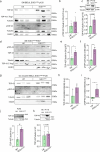
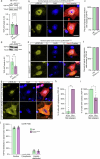
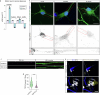
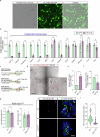
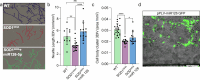
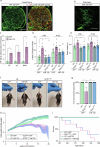
Amyotrophic lateral sclerosis (ALS) is characterized by neuromuscular junction (NMJ) disruption and neurodegeneration. Recent findings highlight a pivotal role for TAR DNA-binding protein 43 (TDP-43) in forming axonal pathological condensates and facilitating NMJ disruption through inhibition of local protein synthesis. However, the mechanisms that drive local TDP-43 accumulation remain unknown. Here we identify that the TDP-43 axonal accumulation in peripheral nerves of SOD1 patients and mice stems from its aberrant local synthesis. This is a non-cell-autonomous process driven by muscle-derived miR-126a-5p extracellular vesicles (EVs). Inhibiting muscle secretion of miR-126a-5p prompts presynaptic TDP-43 synthesis and accumulation, which disrupts axonal translation and causes NMJ degeneration. Introducing miR-126 to SOD1G93A mice, primary co-cultures and human induced pluripotent stem cell (iPSC)-derived co-cultures with ALS mutations exhibits neuroprotective effects and delays motor decline. These findings identify a transcellular communication axis between muscles and motor neurons that regulates axonal local synthesis and NMJ maintenance, offering insights into ALS onset and progression.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Fischer, L. R. et al. Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp. Neurol.185, 232–240 (2004). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

