A circRNA promotes esophageal squamous cell carcinoma progression by inhibiting TRIM25-mediated degradation of IGF2BP family members
- PMID: 41044609
- PMCID: PMC12495880
- DOI: 10.1186/s12943-025-02442-3
A circRNA promotes esophageal squamous cell carcinoma progression by inhibiting TRIM25-mediated degradation of IGF2BP family members
Abstract
Background: Esophageal squamous cell carcinoma (ESCC) is a highly aggressive malignancy with poor prognosis and limited treatment options. While circular RNAs (circRNAs) are frequently dysregulated in ESCC, their functional roles and molecular mechanisms in tumor progression remain largely unexplored.
Methods: We characterized circRNA using RT-PCR, Sanger sequencing, and fluorescent in situ hybridization. Gain- and loss-of-function studies in vitro and in vivo were performed to assess circRNA function. Molecular interactions were investigated via RNA pull-down assays coupled with mass spectrometry, electrophoretic mobility shift assays, co-immunoprecipitation, and immunoblot analysis. Clinical relevance was evaluated by RT-qPCR and immunohistochemistry in patient specimens.
Results: We identified circLNF, a novel circRNA derived from the long non-coding RNA FIRRE gene, which is significantly upregulated in ESCC. Functional assays demonstrated that circLNF promotes proliferative and migratory capacities in cultured cells and accelerates tumor progression and metastasis in animal models. Mechanistically, circLNF directly interacts with IGF2BP family proteins through their "CAUC" motifs, protecting them from TRIM25-mediated ubiquitination and proteasomal degradation. Importantly, circLNF expression positively correlated with IGF2BP1 protein levels in ESCC patient tissues, underscoring the clinical relevance of the circLNF-IGF2BP axis in ESCC progression.
Conclusions: Our findings reveal an oncogenic role of circLNF in ESCC by inhibiting TRIM25-mediated proteasomal degradation of IGF2BP proteins and highlight circLNF as a potential therapeutic target for ESCC.
Keywords: Esophageal squamous cell carcinoma; IGF2BPs; TRIM25; circRNA.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All mouse procedures were approved by the Institutional Animal Care and Use of West China Hospital, Sichuan University (No. 20230227073). Human ESCC samples were obtained from West China Hospital, Sichuan University, with approval from the hospital’s Ethics Committee (No. 2023 − 1764). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




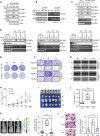

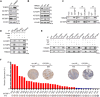
References
-
- Song Y, Li L, Ou Y, Gao Z, Li E, Li X, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–5. - PubMed
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. - PubMed
-
- Liu CX, Chen LL. Circular rnas: characterization, cellular roles, and applications. Cell. 2022;185:2016–34. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

