Cryo-EM studies of amyloid-β fibrils from human and murine brains carrying the Uppsala APP mutation (Δ690-695)
- PMID: 41044681
- PMCID: PMC12492897
- DOI: 10.1186/s40478-025-02120-x
Cryo-EM studies of amyloid-β fibrils from human and murine brains carrying the Uppsala APP mutation (Δ690-695)
Abstract
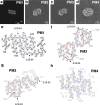
Today, 13 intra-amyloid-β (Aβ) amyloid precursor protein (APP) gene mutations are known to cause familial Alzheimer's disease (AD). Most of them are point mutations causing an increased production or a change in the conformation of Aβ. The Uppsala APP mutation (Δ690-695 in APP, Δ19-24 in Aβ) is the first known multi-codon deletion causing autosomal dominant AD. Here, we applied cryo-electron microscopy (cryo-EM) to investigate the structure of Aβ fibrils with the Uppsala APP mutation from tg-UppSwe mouse brain tissue. Murine AβUpp(1-42)Δ19-24 are made of two identical S-shaped protofilaments with an ordered fibril core of S8-A42. The murine Aβ fold is almost identical to previously described human type II filaments, although the amino acid sequences differ considerably. In addition, we report the cryo-EM structure of Aβ fibrils from the temporal cortex of a patient with the Uppsala APP mutation. The observed structure of the human Aβ fold closely resembles previously described type I fibrils. Structural modeling suggests that these fibrils are composed of wild-type Aβ, which implies that AβUpp may be less soluble and thus not readily accessible for cryo-EM image processing and structure determination. Additionally, from the human sample we determined the structures of tau paired helical filaments and tau straight filaments, which are identical to those found in sporadic AD cases. Finally, we present the 3D cryo-EM structures of four dominant AβUpp(1-42)Δ19-24 fibril polymorphs, formed in vitro. All four polymorphs differ from the observed folds of Uppsala Aβ in murine and human brain tissue, respectively.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Breeding and methods for brain isolation were approved by the Uppsala County Animal Ethics boards (5.8.18-20401/20), following the rules and regulations of the Swedish Animal Welfare Agency, and were in compliance with the European Communities Council Directive of 22 September 2010 (2010/63/EU). The collection and study of the human APP Upp brain were approved by the Uppsala Regional Ethical Review Board (2005-103) and The Swedish Ethical Review Authority (2021-04356), respectively. Consent for publication: Not applicable. Competing interests: MI is a paid consultant to BioArctic AB and Eisai Pharmaceuticals. DW is a founder and shareholder of the companies Priavoid and Attyloid and a member of their supervisory boards. This did not influence the interpretation of the data. All other authors declare no competing interests.
Figures



References
-
- Arakhamia T, Lee CE, Carlomagno Y, Kumar M, Duong DM, Wesseling H, Kundinger SR, Wang K, Williams D, DeTure M, Dickson DW, Cook CN, Seyfried NT, Petrucelli L, Steen JA, Fitzpatrick AWP (2020) Posttranslational modifications mediate the structural diversity of tauopathy strains. Cell 180:633-644.e12. 10.1016/j.cell.2020.01.027 - PMC - PubMed
-
- Bugiani O, Giaccone G, Rossi G, Mangieri M, Capobianco R, Morbin M, Mazzoleni G, Cupidi C, Marcon G, Giovagnoli A, Bizzi A, Di Fede G, Puoti G, Carella F, Salmaggi A, Romorini A, Patruno GM, Magoni M, Padovani A, Tagliavini F (2010) Hereditary cerebral hemorrhage with amyloidosis associated with the E693K mutation of APP. Arch Neurol. 10.1001/archneurol.2010.178 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

