A central research portal for mining pancreatic clinical and molecular datasets and accessing biobanked samples
- PMID: 41045640
- PMCID: PMC12523802
- DOI: 10.1016/j.tranon.2025.102550
A central research portal for mining pancreatic clinical and molecular datasets and accessing biobanked samples
Abstract
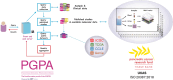
Objectives: We present Pancreas Genome Phenome Atlas (PGPA) as a resource for the mining and analysis of pancreatic -omics datasets, and demonstrate the biological interpretations possible due to this dynamic analytics hub accommodating an extensive range of publicly available datasets.
Methods: Clinical and molecular datasets from four primary sources are included (The Cancer Genome Atlas, International Cancer Genome Consortium, Cancer Cell Line Encyclopaedia, Genomics Evidence Neoplasia Information Exchange), which form the foundation of -omics profiling of pancreatic malignancies and related lesions (n = 7760 specimens). Several user-friendly analytical tools to integrate and explore molecular data derived from these primary specimens and cell lines are available. Crucially, PGPA is positioned as the data access point for Pancreatic Cancer Research Fund Tissue Bank - the only national pancreatic cancer biobank in the UK. This will pioneer a new era of biobanking to promote collaborative studies and effective sharing of multi-modal molecular, histopathology and imaging data (>125 000 specimens from >3980 cases and controls; >2700 radiology images, and >2630 digitised H&Es from 401 donors) to accelerate validation of in silico findings in patient-derived material.
Results: We demonstrate the practical utility of PGPA by investigating somatic variants associated with established transcriptomic subtypes and disease prognosis: several patient-specific variants are clinically actionable and may be leveraged for precision medicine.
Conclusions: This places PGPA at the analytical forefront of pancreatic biomarker-based research, providing the user community with a distinct resource to facilitate hypothesis-testing on public data, validate novel research findings, and access curated, high-quality patient tissues for translational research.
Keywords: Biomarkers; Genomics; Pancreas biobank; Transcriptomics; Translational.
Copyright © 2025. Published by Elsevier Inc.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
LinkOut - more resources
Full Text Sources

