BC-predict: mining of signal biomarkers and production of models for early-stage breast cancer subtyping and prognosis
- PMID: 41048341
- PMCID: PMC12488574
- DOI: 10.3389/fbinf.2025.1644695
BC-predict: mining of signal biomarkers and production of models for early-stage breast cancer subtyping and prognosis
Abstract
Introduction: Disease heterogeneity is the hallmark of breast cancer, which is the most common female malignancy. With a disturbing increase in mortality and disease burden, there remains a need for effective early-stage theragnostic and prognostic biomarkers. In this work, we improved on BrcaDx (https://apalania.shinyapps.io/brcadx/) for cancer vs control screening and examined a cluster of adjoining learning problems in breast cancer heterogeneity: (i) identification of metastatic cancers; (ii) molecular subtyping (TNBC, HER2, or luminal); and (iii) histological subtyping (invasive ductal or invasive lobular).
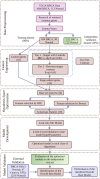
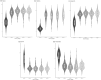
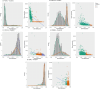
Methods: We analyzed the transcriptomic profiles of breast cancer patients from public-domain databases such as the TCGA using stage-encoded problem-specific statistical models of gene expression and unveiled stage-salient and progression-significant genes. Using a consensus approach, we identified potential machine learning features, and considered six model classes for each learning problem, with hyperparameter optimization on a training dataset and evaluation on a holdout test dataset. A nested approach enabled us to identify the best model class for each learning problem.
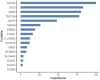
Results: External validation of the best models yielded balanced accuracies of 97.42% for cancer vs normal; 88.22% for metastatic v/s non metastatic; 88.79% for ternary molecular subtyping; and ensemble accuracy of 94.23% for histological subtyping. The model for molecular subtyping was validated on a 26-sample TNBC-only out-of-distribution cohort, yielding 25 correct predictions. We performed a late integration of multi-omics datasets by validating the feature space used in each problem with miRNA profiles, methylation profiles, and commercial breast cancer panels.
Discussion: Pending prospective studies, we have translated the models into BC-Predict that forks the best models developed for each problem in a unified interface and provides a complete readout for input instances of expression data, including uncertainty estimates. BC-Predict is freely available for non-commercial purposes at: https://apalania.shinyapps.io/BC-Predict.
Keywords: biomarker signature discovery; breast cancer heterogeneity; explainable AI; integrative multi-omics; machine learning; metastatic disease; molecular and histological subtype; stage-specific differential gene expression.
Copyright © 2025 Muthamilselvan, Vaithilingam and Palaniappan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Associated data
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

