MicroRNA mapping of bronchial aspirate for molecular phenotyping and prognostication in patients on mechanical ventilation
- PMID: 41049088
- PMCID: PMC12495064
- DOI: 10.1016/j.omtn.2025.102714
MicroRNA mapping of bronchial aspirate for molecular phenotyping and prognostication in patients on mechanical ventilation
Abstract
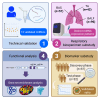
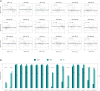
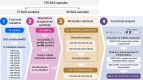
The application of microRNA (miRNA) profiling in respiratory biospecimens, particularly bronchial aspirate (BAS), remains underexplored. Here, we aimed to validate and refine miRNA quantification in BAS samples to establish its suitability for molecular phenotyping. This was a multicenter study including 288 COVID-19 patients on invasive mechanical ventilation. Respiratory biospecimens included BAS, tracheal aspirate, and bronchoalveolar lavage fluid samples. A predesigned miRNA panel was evaluated using RT-qPCR. Biomarker evaluation and functional assessment were subsequently conducted. An initial technical validation phase corroborated the reproducibility of miRNA profiling in BAS samples. Comparative analyses of miRNA expression profiles across respiratory samples revealed distinct miRNA patterns among biospecimens. In the biomarker analysis, two miRNA ratios, miR-34c-5p/miR-34a-5p and miR-34c-5p/miR-125b-5p, were inversely associated with intensive care unit (ICU) survival (hazard ratio [HR]: 0.18 and 0.17, respectively) during the discovery phase. Risk and survival analyses in the test phase confirmed the reproducibility of the miR-34c-5p/miR-34a-5p ratio (hazard ratio [HR] = 0.17). Functional analyses revealed the utility of miRNA profiling in BAS for identifying pathogenic pathways and developing therapeutic targets. Overall, these findings position miRNA profiling in BAS samples as a valuable approach for biomarker discovery, identification of pathophysiological mechanisms, and development of targeted pulmonary therapies.
Keywords: COVID-19; MT: Non-coding RNAs; SARS-CoV-2; biomarkers; bronchial aspirate; bronchoalveolar lavage fluid; invasive mechanical ventilation; microRNAs; noncoding RNA; tracheal aspirate.
© 2025 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- de Gonzalo-Calvo D., Karaduzovic-Hadziabdic K., Dalgaard L.T., Dieterich C., Perez-Pons M., Hatzigeorgiou A., Devaux Y., Kararigas G. Machine learning for catalysing the integration of noncoding RNA in research and clinical practice. EBioMedicine. 2024;106 doi: 10.1016/j.ebiom.2024.105247. - DOI - PMC - PubMed
-
- Perez-Pons M., Molinero M., Benítez I.D., García-Hidalgo M.C., Chatterjee S., Bär C., González J., Torres A., Barbé F., de Gonzalo-Calvo D. MicroRNA-centered theranostics for pulmoprotection in critical COVID-19. Mol. Ther. Nucleic Acids. 2024;35 doi: 10.1016/J.OMTN.2024.102118. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

