Adaptive Potential of Syzygium maire, a Critically Threatened Habitat Specialist Tree Species in Aotearoa New Zealand
- PMID: 41049769
- PMCID: PMC12489745
- DOI: 10.1111/eva.70161
Adaptive Potential of Syzygium maire, a Critically Threatened Habitat Specialist Tree Species in Aotearoa New Zealand
Abstract
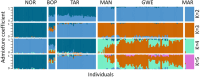
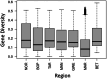
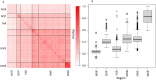
The restoration of swampland is vital for the recovery of both biodiversity and cultural values in Aotearoa New Zealand. Syzygium maire, an endemic wetland tree species, is a focus of many wetland restoration efforts. Formerly widespread, extant populations are small, fragmented, and under pressure from myrtle rust. Restoration initiatives may be unknowingly compounding these threats to the species by failing to represent the complete genetic diversity of populations. What genetic diversity remains in remnants and how it is distributed is not known. We therefore aimed to assess the national scale population structure, genetic diversity, and adaptive potential of S. maire to inform species conservation. We identified over 760,000 high-quality single nucleotide variants in 269 reproductive age trees from across the species' range, using low coverage whole genome resequencing. At a national scale, we found five distinct regional-scale genetic clusters, which in turn exhibit local structure and admixture. In the North Island: Northland, Bay of Plenty in the central east, Taranaki in the central west, and Greater Wellington/Manawatū in the south. A single cluster was identified in the South Island, Marlborough. Within-cluster substructure was particularly evident for Greater Wellington/Manawatū. Genetic diversity and fixation indices (F ST) were relatively uniform across all clusters, and there was some evidence of north to south increase in kinship and shorter time since radiation. These patterns are likely to reflect glaciation cycles that resulted in complex contractions into local microrefugia and subsequent re-radiations of the species over time. Genotype by environment analysis detected genetic variants potentially contributing to environmental adaptation, notably precipitation seasonality. Restoration and conservation goals would best be served by capturing diversity within regional clusters. Information on the geographic and environmentally structured distribution of this tree's genetic diversity supports conservation and restoration strategies through ensuring the complete extant diversity is captured, identifying regions at most risk of genetic degradation, and facilitating planning regarding the movement of adaptive diversity in a changing environment.
© 2025 The Author(s). Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Anderson, S. H. , Kelly D., Ladley J. J., Molloy S., and Terry J.. 2011. “Cascading Effects of Bird Functional Extinction Reduce Pollination and Plant Density.” Science 331: 1068–1071. - PubMed
-
- Andrello, M. , Manel S., Vilcot M., Xuereb A., and D'Aloia C. C.. 2023. “Benefits of Genetic Data for Spatial Conservation Planning in Coastal Habitats.” Cambridge Prisms: Coastal Futures 1: e28.
-
- Andrews, S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Bioinformatics, Babraham Institute.
LinkOut - more resources
Full Text Sources

