Hypoxia-induced RCOR2 promotes macrophage M2 polarization and CD8+ T-cell exhaustion by enhancing LIF transcription in hepatocellular carcinoma
- PMID: 41052883
- PMCID: PMC12506451
- DOI: 10.1136/jitc-2025-012314
Hypoxia-induced RCOR2 promotes macrophage M2 polarization and CD8+ T-cell exhaustion by enhancing LIF transcription in hepatocellular carcinoma
Abstract
Background: The hypoxic microenvironment plays a crucial role in regulating the progression of hepatocellular carcinoma (HCC) and facilitating immune evasion. It is essential to gain a more comprehensive understanding of the pathways through which hypoxia influences HCC progression and immune evasion.
Methods: We employed RNA sequencing, The Cancer Genome Atlas (TCGA) data analysis, clinical data analysis of HCC, and tissue microarray immunohistochemical analysis to identify key genes associated with hypoxia regulation and immune evasion. We investigated the biological functions of REST corepressor 2 (RCOR2) in tumor progression and immune evasion through mass cytometry, multiplex immunofluorescence, an orthotopic liver transplantation tumor model, in vitro co-culture systems, flow cytometry, and immunohistochemical analysis. Additionally, we used molecular techniques such as RNA sequencing, chromatin immunoprecipitation sequencing, and mass spectrometry to gain deeper insights into the potential molecular mechanisms underlying RCOR2.
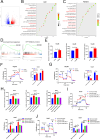
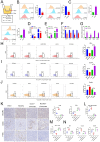
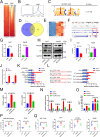
Results: We found that the hypoxia-related factor RCOR2 is upregulated in HCC and is associated with a poor prognosis. RCOR2 enhances the glycolytic process in HCC cells, thereby promoting the proliferation and metastasis of HCC cells under hypoxic conditions. Additionally, RCOR2 facilitates the M2 polarization of macrophages and contributes to the exhaustion of CD8+ T cells. Mechanistically, the hypoxic microenvironment increases the expression of RCOR2 through hypoxia-inducible factor 1-alpha. Concurrently, this microenvironment inhibits the ubiquitin-mediated degradation of RCOR2 by promoting its sumoylation, which facilitates its translocation to the nucleus. The sumoylation of RCOR2 further enhances the transcriptional activity of leukemia inhibitory factor (LIF). LIF, derived from HCC, contributes to the M2 polarization of macrophages, thereby facilitating immune evasion and playing a role in resistance to programmed cell death protein 1 (PD-1) therapies.
Conclusions: Our research reveals that the RCOR2/LIF axis within the hypoxic microenvironment of HCC plays a significant role in immune evasion and identifies novel biomarkers associated with tumor resistance to anti-PD-1 therapy. This study provides potential therapeutic targets for HCC.
Keywords: Hepatocellular Carcinoma; Immunotherapy; Macrophage.
© Author(s) (or their employer(s)) 2025. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ Group.
Conflict of interest statement
Competing interests: No, there are no competing interests.
Figures






References
-
- Koukourakis MI, Giatromanolaki A, Skarlatos J, et al. Hypoxia inducible factor (HIF-1a and HIF-2a) expression in early esophageal cancer and response to photodynamic therapy and radiotherapy. Cancer Res. 2001;61:1830–2. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
