Virulence characteristics and antibiotic resistance analysis of multi-drug resistant Enterotoxigenic Escherichia coli in healthy pigs from Zhejiang Province, China
- PMID: 41053715
- PMCID: PMC12502167
- DOI: 10.1186/s12917-025-05044-9
Virulence characteristics and antibiotic resistance analysis of multi-drug resistant Enterotoxigenic Escherichia coli in healthy pigs from Zhejiang Province, China
Abstract
Background: Enterotoxigenic Escherichia coli (ETEC) is a predominant diarrheal pathogen worldwide, employing adhesins and secreted enterotoxins to cause disease in mammalian hosts. The report about antimicrobial resistance (AMR) in ETEC have increased, detrimentally affecting both livestock industry and food safety. Despite this, there is a paucity of studies on the AMR genes, virulence factors and the correlation of them in ETEC.
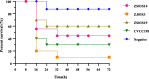
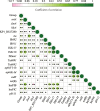
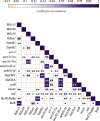
Results: Six ETEC-positive samples collected from healthy pigs across three regions yielded 16 distinct ETEC isolates. Among these, 13 isolates (81.3%) exhibited multidrug resistance (MDR), while all 16 isolates demonstrated tolerance to working concentrations of sodium hypochlorite. 15 isolates harbored both heat-labile enterotoxin genes eltAB and heat-stable enterotoxin gene stb. One isolate uniquely carried eltAB, heat-stable enterotoxin genes sta, stb, and Shiga toxin gene stx2e. Galleria mellonella infection assays confirmed the virulence of representative strains in our study. Whole-genome sequencing of one isolate per sample (n = 6), integrated with NCBI porcine ETEC data, revealed broad distribution of sequence types (STs). A previously unreported ST7070 strain was identified as an enterotoxin gene carrier (eltAB + stb). The results of correlation analysis revealed that co-transfer in aminoglycoside resistance genes aph(3'')-Ib, aph(6')-Id, sulfonamide resistance gene sulIII and co-localized cluster of arr-3 (rifampicin resistance), aac(6')-Ib-cr (aminoglycoside resistance), catB (phenicol resistance), and blaOXA (β-lactam resistance). Significant co-transfer observed between disinfectant resistance gene sugE(p) and extended-spectrum β-lactamase gene blaCMY.
Conclusions: ETEC strains isolated from healthy animals exhibited both pathogenicity and MDR phenotypes. Besides, they harbored diverse AMR genes and disinfectant resistance determinants. Integrated analysis with NCBI-deposited ETEC genomic data further revealed co-transfer events of AMR and disinfectant resistance genes among ETEC populations.
Supplementary Information: The online version contains supplementary material available at 10.1186/s12917-025-05044-9.
Keywords: Antimicrobial resistance; Bioinformatic analysis; ETEC; Pigs.
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All sample collections in this study were conducted with the full knowledge and consent of the owners of pig farms. This study was approved by the Institutional Animal Care and Use Committee of the Science Technology Department of Zhejiang Province in accordance with the Regulations for the Administration of Affairs Concerning Experimental Animals. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures






References
-
- Laird TJ, Abraham S, Jordan D, et al. Porcine enterotoxigenic Escherichia coli: antimicrobial resistance and development of microbial-based alternative control strategies. Vet Microbiol. 2021;258:109117. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

