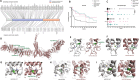Mutations in the Key Autophagy Tethering Factor EPG5 Link Neurodevelopmental and Neurodegenerative Disorders Including Early-Onset Parkinsonism
- PMID: 41053928
- PMCID: PMC12577676
- DOI: 10.1002/ana.78013
Mutations in the Key Autophagy Tethering Factor EPG5 Link Neurodevelopmental and Neurodegenerative Disorders Including Early-Onset Parkinsonism
Abstract
Objective: Autophagy is a fundamental biological pathway with vital roles in intracellular homeostasis. During autophagy, defective cargoes including mitochondria are targeted to lysosomes for clearance and recycling. Recessive truncating variants in the autophagy gene EPG5 have been associated with Vici syndrome, a severe early-onset neurodevelopmental disorder with extensive multisystem involvement. Here, we aimed to delineate the extended, age-dependent EPG5-related disease spectrum.
Methods: We investigated clinical, radiological, and molecular features from the largest cohort of EPG5-related patients identified to date, complemented by experimental investigation of cellular and animal models of EPG5 defects.
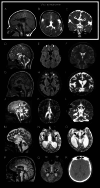
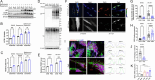
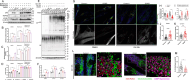
Results: Through worldwide collaboration, we identified 211 patients, 97 of them previously unpublished, with recessive EPG5 variants. The phenotypic spectrum ranged from antenatally lethal presentations to milder isolated neurodevelopmental disorders. A novel Epg5 knock-in mouse model of a recurrent EPG5 missense variant featured motor impairments and defective autophagy in brain areas particularly relevant for the neurological disorders in milder presentations. Novel age-dependent neurodegenerative manifestations in our cohort included adolescent-onset parkinsonism and dystonia with cognitive decline, and myoclonus. Radiological features suggested an emerging continuum with brain iron accumulation disorders. Patient fibroblasts showed defects in PINK1-Parkin-dependent mitophagic clearance and α-synuclein overexpression, indicating a cellular basis for the observed neurodegenerative phenotypes. In Caenorhabditis elegans, EPG5 knockdown caused motor impairments, defective mitophagic clearance, and changes in mitochondrial respiration comparable to observations in C. elegans knockdown of parkinsonism-related genes.
Interpretation: Our findings illustrate a lifetime neurological disease continuum associated with pathogenic EPG5 variants, linking neurodevelopmental and neurodegenerative disorders through the common denominator of defective autophagy. ANN NEUROL 2025;98:932-950.
© 2025 The Author(s). Annals of Neurology published by Wiley Periodicals LLC on behalf of American Neurological Association.
Conflict of interest statement
Nothing to report.
Figures