Logical Exploration of Cinnamoyl-Containing Nonribosomal Peptides via Metabologenomic Targeting and Regulator Overexpression
- PMID: 41055158
- PMCID: PMC12532209
- DOI: 10.1021/jacs.5c13143
Logical Exploration of Cinnamoyl-Containing Nonribosomal Peptides via Metabologenomic Targeting and Regulator Overexpression
Abstract
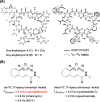
A targeted method for discovering cinnamoyl-containing nonribosomal peptides (CCNPs), a unique class of bioactive compounds, was devised by using cinnamoyl isomerase, a key enzyme in the biosynthesis of the cinnamoyl moiety, as a genome mining probe. A total of 39 hit strains were obtained, including 35 from polymerase chain reaction-based screening of the in-house bacterial library (2.5% of 1400 strains) targeting the cinnamoyl isomerase-encoding gene and 4 from the genome mining of online databases. Sequence similarity networking and phylogenetic analyses of the isomerase amplicons (∼530 bp) classified the CCNPs into three major substructure-based groups (Z-, E-, and M-type CCNPs) and revealed distinct clade-structure relationships (13 clades). To overcome the challenge of silent biosynthetic gene clusters, we activated these clusters by overexpressing conserved cluster-situated LuxR regulators combined with extensive culture optimization. CCNP production was metabolomically detected in the bacterial extracts by using the characteristic UV absorption and MS/MS fragments of cinnamoyl moieties. CCNP production was observed in 20 of the 39 hit strains, resulting in the isolation of 6 new CCNPs, including oxy-skyllamycin B (2), gwanacinnamycin (3), and luxocinnamycins A-D (4-7), with high structural novelty. Their structures were elucidated using comprehensive spectroscopic analyses and multiple-step chemical derivatizations, and the putative biosynthetic pathways were bioinformatically proposed. Gwanacinnamycin (3) exhibited significant antimycobacterial activity, whereas luxocinnamycin A (4) displayed moderate antiproliferative activity against stomach cancer cells. Our findings highlight a targeted metabologenomic approach combined with transcriptional regulator overexpression as a logical and efficient platform for the discovery of bioactive compounds from nature.
Figures





References
-
- Hayashi K., Hashimoto M., Shigematsu N., Nishikawa M., Ezaki M., Yamashita M., Kiyoto S., Okuhara M., Kohsaka M., Imanaka H.. WS9326A, a Novel Tachykinin Antagonist Isolated from Streptomyces violaceusniger No. 9326. J. Antibiot. 1992;45(7):1055–1063. doi: 10.7164/antibiotics.45.1055. - DOI - PubMed
-
- Toki S., Agatsuma T., Ochiai K., Saitoh Y., Ando K., Nakanishi S., Lokker N. A., Giese N. A., Matsuda Y.. RP-1776, a Novel Cyclic Peptide Produced by Streptomyces sp., Inhibits the Binding of PDGF to the Extracellular Domain of Its Receptor. J. Antibiot. 2001;54(5):405–414. doi: 10.7164/antibiotics.54.405. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

