Immunophenotypic changes in the tumor and tumor microenvironment during progression to multiple myeloma
- PMID: 41056512
- PMCID: PMC12558612
- DOI: 10.1371/journal.pgen.1011848
Immunophenotypic changes in the tumor and tumor microenvironment during progression to multiple myeloma
Abstract
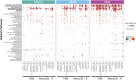
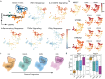
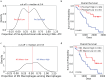
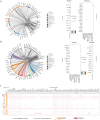
Investigation of the cellular and molecular mechanisms of disease progression from precursor plasma cell disorders to active disease increases our understanding of multiple myeloma (MM) pathogenesis and supports the development of novel therapeutic strategies. In this analysis, single-cell RNA sequencing, surface protein profiling, and B lymphocyte antigen receptor profiling of unsorted, whole bone marrow (BM) mononuclear cell samples was used to study molecular changes in tumor cells and the tumor microenvironment (TME). A cell atlas of the BM microenvironment was generated from 123 subjects including healthy volunteers and patients with monoclonal gammopathy of unknown significance (MGUS), smoldering MM (SMM), and MM. These analyses revealed commonalities in molecular pathways, including MYC signaling, E2F targets and interferon alpha response, that were altered during disease progression. Evidence of early dysregulation of the immune system in MGUS and SMM, which increases and impacts many cell types as the disease progresses, was found. In parallel with disease progression, population shifts in CD8 + T cells, macrophages, and classical dendritic cells were observed, and the resulting differences in CD8 + T cells and macrophages were associated with poor overall survival outcomes. Potential ligand-receptor interactions that may play a role during the transition from precursor stages to MM were identified, along with potential biomarkers of disease progression, some of which may represent novel therapeutic targets. MIF, IL15, CD320, HGF and FAM3C were detected as potential regulators of the TME by plasma cells, while SERPINA1 and BAFF (TNFSF13B) were found to have the highest potential to contribute to the downstream changes observed between precursor stage and MM cells. These findings demonstrate that myeloma tumorigenesis is associated with dysregulation of molecular pathways driven by gradually occurring immunophenotypic changes in the tumor and TME. Trial registration: This project has been registered at EudraCT (European Union Drug Regulating Authorities Clinical Trials Database) with protocol number NOPRODMMY0001 and EudraCT Number 2018-004443-23 on 12 December 2018.
Copyright: © 2025 Bergiers et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: IB, SS, NF, GV, TS, BV, DDM, JVH, KVdB, RV, BH, CJH, and TC were employees of Janssen Research & Development LLC at the time the study was conducted and may hold stock and/or stock options. MCK and NB received research support from Janssen Research & Development LLC. JC has a research mandate funded by the Fondation contre le Cancer. MM, J-CE, WK, MD, NM, JVD, PV and YB have declared that no competing interests exist.
Figures




References
-
- National Cancer Institute (NCI). Surveillance, Epidemiology, and End Results (SEER) Program. Available from: https://seer.cancer.gov/
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

