Methylation reprogramming associated with aggressive prostate cancer and ancestral disparities
- PMID: 41057544
- PMCID: PMC12673094
- DOI: 10.1038/s44320-025-00153-x
Methylation reprogramming associated with aggressive prostate cancer and ancestral disparities
Abstract
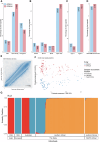
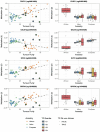
African men are disproportionately impacted by aggressive prostate cancer (PCa). The key to this disparity is both genetic and environmental factors, alluding to epigenetic modifications. However, African-inclusive prostate tumour DNA methylation studies are lacking. Assembling a multi-geo-ancestral prostate tissue cohort, including men with (57 African, 48 European, 23 Asian) or without (65 African) PCa, we interrogate for genome-wide differential methylation. Overall, methylation appears to be driven by ancestry over geography (152 southern Africa, 41 Australia). African tumours show substantial heterogeneity, with universal hypermethylation indicating more pervasive epigenetic silencing, encompassing PCa suppressor genes and enhancer-targeted binding motifs. Conversely, African tumour-associated heterochromatic hypomethylation suggests chromatin relaxation and developmental pathway activation via enhancer targets. Notably, non-prostate lineage elements appeared preferentially exploited in African tumorigenesis, with ancestry potentially influencing the extent of lineage-inappropriate activation, and tumour progression marked by repression of developmental regulators. Together, these findings point to extensive epigenetic plasticity in African tumours, with intergenic regulatory remodelling promoting genomic instability, metastatic potential and aggressive disease phenotypes.
Keywords: African Ancestry; Differential Methylation; Health Disparity; Prostate Tumours.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. Hayes is a Member of Active Surveillance Movember Committee and received an honorarium from The Korean Urological Oncology Society for 2024 Annual Conference as a guest speaker.
Figures






Update of
-
Methylation reprogramming associated with aggressive prostate cancer and ancestral disparities.bioRxiv [Preprint]. 2025 Apr 16:2025.04.16.649098. doi: 10.1101/2025.04.16.649098. bioRxiv. 2025. Update in: Mol Syst Biol. 2025 Dec;21(12):1676-1701. doi: 10.1038/s44320-025-00153-x. PMID: 40321218 Free PMC article. Updated. Preprint.
References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, Jemal A (2024) Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 74:229–263 - PubMed
MeSH terms
Grants and funding
- PC210168,PC230673/DOD | MHS | Congressionally Directed Medical Research Programs (CDMRP)
- R01 CA285772/CA/NCI NIH HHS/United States
- PC200390/DOD | MHS | Congressionally Directed Medical Research Programs (CDMRP)
- 1R01CA285772-01/HHS | NIH | NCI | Center for Cancer Research (CCR)
- PMDS22070633683/DSI | National Research Foundation (NRF)
LinkOut - more resources
Full Text Sources
Medical

