Neutrophil extracellular trapping network-associated biomarkers in liver fibrosis: machine learning and experimental validation
- PMID: 41057887
- PMCID: PMC12502331
- DOI: 10.1186/s12967-025-06819-2
Neutrophil extracellular trapping network-associated biomarkers in liver fibrosis: machine learning and experimental validation
Abstract
Background: The diagnostic and therapeutic potential of neutrophil extracellular traps (NETs) in liver fibrosis (LF) has not been fully explored. We aim to screen and verify NETs-related liver fibrosis biomarkers through machine learning.
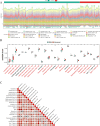
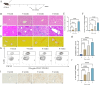
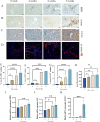
Methods: In order to obtain NETs-related differentially expressed genes (NETs-DEGs), differential analysis and WGCNA analysis were performed on the GEO dataset (GSE84044, GSE49541) and the NETs dataset. Enrichment analysis and protein interaction analysis were used to reveal the candidate genes and potential mechanisms of NETs-related liver fibrosis. Biomarkers were screened using SVM-RFE and Boruta machine learning algorithms, followed by immune infiltration analysis. A multi-stage model of fibrosis in mice was constructed, and neutrophil infiltration, NETs accumulation and NETs-related biomarkers were characterized by immunohistochemistry, immunofluorescence, flow cytometry and qPCR. Finally, the molecular regulatory network and potential drugs of biomarkers were predicted.
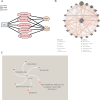
Results: A total of 166 NETs-DEGs were identified. Through enrichment analysis, these genes were mainly enriched in chemokine signaling pathway and cytokine-cytokine receptor interaction pathway. Machine learning screened CCL2 as a NETs-related liver fibrosis biomarker, involved in ribosome-related processes, cell cycle regulation and allograft rejection pathways. Immune infiltration analysis showed that there were significant differences in 22 immune cell subtypes between fibrotic samples and healthy samples, including neutrophils mainly related to NETs production. The results of in vivo experiments showed that neutrophil infiltration, NETs accumulation and CCL2 level were up-regulated during fibrosis. A total of 5 miRNAs, 2 lncRNAs, 20 function-related genes and 6 potential drugs were identified based on CCL2.
Conclusions: This study identified CCL2 as a biomarker for nets-related liver fibrosis, providing a new perspective for understanding the mechanisms of nets-mediated fibrosis and promoting therapeutic discovery.
Keywords: Bioinformatics; Liver fibrosis; Machine learning; NETs.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Animal experiments for this study were approved by the Institutional Animal Care Committee of Guanganmen Hospital (IACUC-2025-103-SQ). Consent for publication: Not applicable. Competing interests: The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

