Inflammation perturbs hematopoiesis by remodeling specific compartments of the bone marrow niche
- PMID: 41060335
- PMCID: PMC7618457
- DOI: 10.1182/blood.2025029513
Inflammation perturbs hematopoiesis by remodeling specific compartments of the bone marrow niche
Abstract
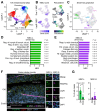
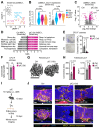
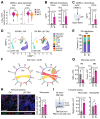
Hematopoietic stem and progenitor cells (HSPC) are regulated by interactions with stromal cells in the bone marrow (BM) cavity, which can be segregated into two spatially defined central marrow (CM) and endosteal (Endo) compartments. However, the importance of this spatial compartmentalization for BM responses to complex conditions like inflammation remains largely unknown. Here, we extensively validate a combination of scRNA-seq profiling and matching flow cytometry isolation that reproducibly identifies 7 key CM and Endo populations and accurately surveys both niche locations. We demonstrate that inflammatory perturbations exert specific effects on different cellular compartments, with type I interferon responses causing leptin receptor-expressing mesenchymal stromal cells to abandon their normal stromal functions and instead adopt an inflammatory phenotype associated with overproduction of chemokines that modulate local monocyte dynamics in the surrounding microenvironment. Our results provide a comprehensive method for molecular and functional stromal characterization and highlight the importance of altered stomal cell activity in regulating hematopoietic responses to inflammatory challenges.
Copyright © 2025 American Society of Hematology.
Conflict of interest statement
Figures




Update of
-
Inflammation perturbs hematopoiesis by remodeling specific compartments of the bone marrow niche.bioRxiv [Preprint]. 2024 Sep 12:2024.09.12.612751. doi: 10.1101/2024.09.12.612751. bioRxiv. 2024. Update in: Blood. 2025 Oct 08:blood.2025029513. doi: 10.1182/blood.2025029513. PMID: 39314376 Free PMC article. Updated. Preprint.
References
-
- Swann JW, Olson OC, Passegué E. Made to order: emergency myelopoiesis and demand-adapted innate immune cell production. Nat Rev Immunol. 2024;24:596–613. - PubMed
-
- Isringhausen S, Mun Y, Kovtonyuk L, Kräutler NJ, Suessbier U, Gomariz A, Spaltro G, Helbling PM, Wong HC, Nagasawa T, Manz MG, et al. Chronic viral infections persistently alter marrow stroma and impair hematopoietic stem cell fitness. J Exp Med. 2021;218:e20192070. doi: 10.1084/jem.20192070. - DOI - PMC - PubMed
-
- Mitchell CA, Verovskaya EV, Calero-Nieto FJ, Olson OC, Swann JW, Wang X, Herault A, Dellorusso PV, Zhang SY, Svendsen AF, Pietras EM, et al. Stromal niche inflammation mediated by IL-1 signalling is a targetable driver of haematopoietic ageing. Nat Cell Biol. 2023;25:30–41. doi: 10.1038/s41556-022-01053-0. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

