Genetic, structural, and functional characterization of allomelanin from black yeast Exophiala viscosa, a chassis for fungal melanin production
- PMID: 41060412
- PMCID: PMC12507995
- DOI: 10.1007/s00253-025-13597-w
Genetic, structural, and functional characterization of allomelanin from black yeast Exophiala viscosa, a chassis for fungal melanin production
Abstract
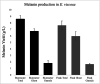
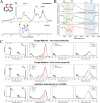
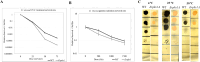
Melanized fungi are known for their remarkable resilience to environmental stress, largely attributed to the protective properties of melanin. In this study, we establish the black yeast Exophiala viscosa as a non-pathogenic, genetically tractable model for the scalable production and functional analysis of DHN-melanin (allomelanin). Cultivation in flasks and bioreactors yielded up to 8.6 g/L of melanin, with the majority tightly incorporated into the cell wall as "melanin ghosts". Chemical analyses including FTIR, XPS, ssNMR, and EPR confirmed the identity of the pigment as allomelanin and revealed a structural association with chitin. Gene deletions of Pks1, Arp2, and Abr2 validated the DHN-melanin biosynthetic pathway and enabled the generation of pigment-deficient mutants. Functional assays demonstrated that melanin contributes significantly to UV and cold tolerance, while offering limited protection against γ-radiation, suggesting that other pigments,such as carotenoids, may also play a protective role. The unique redox properties, structural integrity, and scalability of melanin production in E. viscosa highlight its potential for bio-derived materials used in radiation shielding, environmental remediation, and thermal regulation. This work establishes E. viscosa as a promising chassis for melanin biomanufacturing and a valuable model for studying fungal melanins in the context of materials science and environmental resilience. KEY POINTS: • Cultivation of E. viscosa in rich medium yielded up to 8.6 g/L of melanin. • Chemical and genetic analyses identified the pigment as allomelanin. • Melanin enhanced the tolerance of fungal cells to UV radiation and low temperatures.
Keywords: Exophiala; Allomelanin; Biomanufacturing; DHN-melanin; Melanized fungus; Radiation resistance.
© 2025. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
Declarations. Ethical approval: This article does not contain any studies involving humans or animals conducted by the authors. Competing interests: The authors declare no competing interests.
Figures



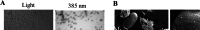
References
-
- Ahmad I, Pawara RH, Girase RT, Pathan AY, Jagatap VR, Desai N, Ayipo YO, Surana SJ, Patel H (2022) Synthesis, molecular modeling study, and quantum-chemical-based investigations of isoindoline-1,3-diones as antimycobacterial agents. ACS Omega 7(25):21820–21844. 10.1021/acsomega.2c01981 - PMC - PubMed
-
- Bell AA, Wheeler MH (1986) Biosynthesis and functions of fungal melanins. Annu Rev Phytopathol 24:411–451. 10.1146/annurev.py.24.090186.002211
-
- Camacho E, Vij R, Chrissian C, Prados-Rosales R, Gil D, O’Meally RN, Cordero RJB, Cole RN, McCaffery JM, Stark RE, Casadevall A (2019) The structural unit of melanin in the cell wall of the fungal pathogen Cryptococcus neoformans. J Biol Chem 294(27):10471–10489. 10.1074/jbc.RA119.008684 - PMC - PubMed
-
- Carr EC, Barton Q, Grambo S, Sullivan M, Renfro CM, Kuo AL, Pangilinan J, Lipzen A, Keymanesh K, Savage E, Barry K, Grigoriev IV, Riekhof WR, Harris SD (2023) Characterization of a novel polyextremotolerant fungus, Exophiala viscosa, with insights into its melanin regulation and ecological niche. G3: Genes Genomes Genetics. 10.1093/g3journal/jkad110 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

