Elucidating molecularly stratified single agent, and combination, therapeutic strategies targeting MCL1 for lethal prostate cancer
- PMID: 41062500
- PMCID: PMC12508096
- DOI: 10.1038/s41467-025-64042-5
Elucidating molecularly stratified single agent, and combination, therapeutic strategies targeting MCL1 for lethal prostate cancer
Abstract
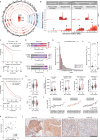
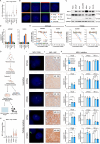
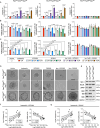
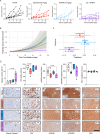
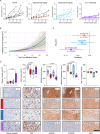
Metastatic castration-resistant prostate cancer (mCRPC) is a lethal disease requiring additional therapeutic strategies. MCL1, an anti-apoptotic BCL2 family member, promotes cancer-cell survival, but its role in mCRPC remains poorly understood. Here, we characterise MCL1 in multiple mCRPC biopsy cohorts and patient-derived models, assessing responses to MCL1 inhibition. MCL1 copy number gain (14%-34%) correlates with increased MCL1 expression and worse outcomes. MCL1 inhibition exhibits anti-tumour effects in MCL1-gained mCRPC models. Co-inhibition of MCL1 and AKT induces cancer-specific cell death in PTEN-loss/PI3K-activated models in vitro and in vivo, modulating BAD-BCLXL and BIM-MCL1 interactions, with durable anti-tumour activity in models with AKT inhibitor acquired resistance. Finally, CDK9-mediated MCL1 downregulation combined with AKT inhibition recapitulates these findings, providing further opportunities for clinical translation. These data support early phase clinical trials targeting MCL1, both as monotherapy for MCL1-gained mCRPC, and in combination with AKT inhibition for PTEN-loss/PI3K-activated mCRPC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.M.J.-V., D.W., I.F., A.d.H.V., A.P., W.Y., G.S., D.B., B.G., C.B., S.M., L.B., W.Z., A.J.N., J.W., J.R., R.P., A.F., M.C., R.R., S.D., J.T., E.H., M.V., J.N., K.L., A.S., F.R., S.C., J.S.d.B. and A.Sharp are employees of the ICR, which has a commercial interest in abiraterone, PARP inhibition in DNA repair defective cancers, and PI3K/AKT pathway inhibitors (no personal income). A.G.S. reports that the National Cancer Institute (NCI) has a Cooperative Research and Development Agreement (CRADA) with Astellas. Resources are provided by this CRADA to the NCI. A.G.S. gets no personal funding from this CRADA but is the primary investigator of the CRADA. J.S.d.B. has served on advisory boards and received fees from many companies, including Amgen, Astra Zeneca, Bayer, Bioxcel Therapeutics, Daiichi, Genentech/Roche, GSK, Merck Serono, Merck Sharp & Dohme, Pfizer, and Sanofi Aventis. He is an employee of the ICR, which has received funding or other support for his research work from AZ, Astellas, Bayer, Cellcentric, Daiichi, Genentech, Genmab, GSK, Janssen, Merck Serono, MSD, Menarini/Silicon Biosystems, Orion, Sanofi Aventis, Sierra Oncology, Taiho, Pfizer, Vertex. J.S.d.B. was named as an inventor, with no financial interest, for patent 8,822,438, submitted by Janssen, that covers the use of abiraterone acetate with corticosteroids. J.S.d.B. has been the CI/PI of many industry-sponsored clinical trials. A.Sharp has received travel support from Sanofi, Roche-Genentech and Nurix, and speaker honoraria from Astellas Pharma and Merck Sharp & Dohme. He has served as an advisor to DE Shaw Research, CHARM Therapeutics, Ellipses Pharma and Droia Ventures. A.Sharp has been the CI/PI of industry-sponsored clinical trials. The remaining authors declare no conflicts of interest.
Figures





References
-
- Sumanasuriya S., De Bono J. Treatment of advanced prostate cancer-a review of current therapies and future promise. Cold Spring Harb. Perspect. Med. 8, a030635 (2018).
-
- Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell144, 646–674 (2011). - PubMed
-
- Westaby, D. et al. Targeting the intrinsic apoptosis pathway: a window of opportunity for prostate cancer. Cancers. 14, 51 (2021).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

