Pathologist-like explainable AI for interpretable Gleason grading in prostate cancer
- PMID: 41062516
- PMCID: PMC12508442
- DOI: 10.1038/s41467-025-64712-4
Pathologist-like explainable AI for interpretable Gleason grading in prostate cancer
Abstract
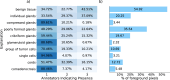
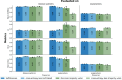
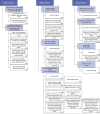
The aggressiveness of prostate cancer is primarily assessed from histopathological data using the Gleason scoring system. Conventional artificial intelligence (AI) approaches can predict Gleason scores, but often lack explainability, which may limit clinical acceptance. Here, we present an alternative, inherently explainable AI that circumvents the need for post-hoc explainability methods. The model was trained on 1,015 tissue microarray core images, annotated with detailed pattern descriptions by 54 international pathologists following standardized guidelines. It uses pathologist-defined terminology and was trained using soft labels to capture data uncertainty. This approach enables robust Gleason pattern segmentation despite high interobserver variability. The model achieved comparable or superior performance to direct Gleason pattern segmentation (Dice score: vs. ) while providing interpretable outputs. We release this dataset to encourage further research on segmentation in medical tasks with high subjectivity and to deepen insights into pathologists' reasoning.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: TJB owns a company that develops mobile apps (Smart Health Heidelberg GmbH, Heidelberg, Germany). TJB received honoraria from Novartis, Roche and HEINE Optotechnik. JNK declares consulting services for Panakeia, AstraZeneca, MultiplexDx, Mindpeak, Owkin, DoMore Diagnostics, and Bioptimus. Furthermore, he holds shares in StratifAI, Synagen, Tremont AI, and Ignition Labs, has received an institutional research grant from GSK, and has received honoraria from AstraZeneca, Bayer, Daiichi Sankyo, Eisai, Janssen, Merck, MSD, BMS, Roche, Pfizer, and Fresenius. YT declares consulting services for Indica Labs, AstraZeneca, MSD, Pfizer; royalties not related to this study (Indica Labs). CMS is a cofounder and shareholder of Vicinity Bio GmbH, and is a scientific advisor to and has received research funding from Enable Medicine Inc., all outside the current work. HM declares consulting services for PathXL, Invicro, and PathAI, outside of this work. NJR discloses an advisory board function for AbbVie AG, and receipt of a travel grant from Roche Diagnostics, both outside of the scope of the current work. IK has received honoraria from AstraZeneca and Menarini Stemline, as well as gifts/financial advantages from Roche (conference invitations). NTG received an institutional research grant from Janssen/Johnson & Johnson and declares consulting services for/honoraria from AstraZeneca, Janssen, Merck, BMS, Daiichi Sankyo, and Bayer. No other conflicts of interest are declared by any of the authors.
Figures





References
-
- Ferlay, J. et al. Cancer statistics for the year 2020: An overview. Int. J. Cancer149, 778–789 (2021). - PubMed
-
- Gleason, D. F., Mellinger, G. T. & Veterans Administration Cooperative Urological Research Group Prediction of Prognosis for Prostatic Adenocarcinoma by Combined Histological Grading and Clinical Staging. J. Urol.197, 134–139 (1974). - PubMed
-
- Epstein, J. I. et al. The 2019 Genitourinary Pathology Society (GUPS) White Paper on Contemporary Grading of Prostate Cancer. Arch. Pathol. Lab. Med.145, 461–493 (2021). - PubMed
-
- Leitlinienprogramm Onkologie (Deutsche Krebsgesellschaft, Deutsche Krebshilfe, AWMF). S3-Leitlinie Prostatakarzinom, Langversion 7.0. AWMF Leitlinienregister. Registernummer 043-022OL. https://www.leitlinienprogramm-onkologie.de/leitlinien/prostatakarzinom/ (2024).

