Superspreading and the evolution of virulence
- PMID: 41066464
- PMCID: PMC12510586
- DOI: 10.1371/journal.pcbi.1013517
Superspreading and the evolution of virulence
Abstract
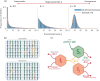
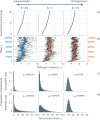
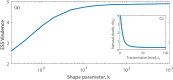
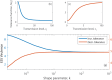
Superspreading, where a small proportion of a population can cause a high proportion of infection transmission, is well known to be important to the epidemiology of a wide range of pathogens, including SARS-CoV-2. However, despite its ubiquity in important human and animal pathogens, the impact of superspreading on the evolution of pathogen virulence is not well understood. Using theory and both deterministic and stochastic simulations we examine the evolution of pathogen virulence under a range of different distributions of infection transmission for the host. Importantly, for many pathogens, superpreader events may be associated with increased tolerance to infection or asymptomatic infection and when we account for this superspreading selects for higher virulence. In contrast, in animal populations where highly connected individuals, that are linked to superspreader events, also have fitness benefits, superspreading may select for milder pathogens. In isolation, the transmission distribution of the host does not impact selection for pathogen virulence. However, superspreading reduces the rate of pathogen evolution and generates considerable variation in pathogen virulence. Therefore, the adaptation of an emerging infectious disease, that exhibits superspreading, is likely to be slowed and characterised by the maintenance of maladaptive variants. Taken as a whole, our results show that superspreading can have important impacts on the evolution of pathogens.
Copyright: © 2025 O’Neill et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

