MUC1 drives ferroptosis resistance in ICC via Src-mediated FSP1 deubiquitination and myristoylation
- PMID: 41068561
- PMCID: PMC12510807
- DOI: 10.1002/ctm2.70495
MUC1 drives ferroptosis resistance in ICC via Src-mediated FSP1 deubiquitination and myristoylation
Abstract
Background: Intrahepatic cholangiocarcinoma (ICC) exhibits poor prognosis and limited therapeutic options. Ferroptosis represents a promising therapeutic strategy, yet resistance mechanisms remain poorly understood. This study investigated the role of mucin 1 (MUC1) in regulating ferroptosis sensitivity in ICC.
Methods: Bioinformatic analyses of GEO and TCGA datasets identified ferroptosis-related factors in ICC. MUC1 expression was validated in ICC cell lines and clinical specimens. Ferroptosis sensitivity was assessed through RSL3-induced cell death assays, lipid peroxidation measurements, and iron detection. Mechanistic studies employed immunoprecipitation-mass spectrometry, co-immunoprecipitation, kinase assays, and deubiquitination assays. In vivo efficacy was evaluated using subcutaneous tumor models.
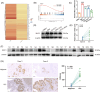
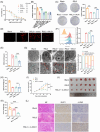
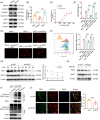
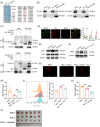
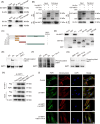
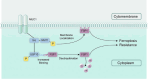
Results: MUC1 was identified as a critical ferroptosis suppressor in ICC. MUC1 overexpression conferred RSL3 resistance by inhibiting lipid peroxidation and reducing ferrous iron accumulation, independent of the GPX4-glutathione pathway. Mechanistically, MUC1 recruited Src kinase, which phosphorylated deubiquitinating enzyme ubiquitin-specific protease 10 (USP10) at tyrosines 359 and 364, enhancing ferroptosis suppressor protein 1 (FSP1) deubiquitination at lysine 246 and stabilizing FSP1 protein. Concurrently, Src phosphorylated N-myristoyltransferase 1 (NMT1) at tyrosine 41, augmenting FSP1 membrane localization through myristoylation. This dual mechanism potentiated the FSP1- coenzyme Q10 (CoQ10) antioxidant system. MUC1 knockdown significantly enhanced ferroptotic sensitivity in vitro and suppressed tumor growth in vivo.
Conclusions: MUC1 orchestrates ferroptosis resistance in ICC through the Src-USP10/NMT1-FSP1 axis. Targeting this signaling cascade represents a potential therapeutic strategy for overcoming ferroptosis resistance in ICC.
Key points: MUC1 suppresses ferroptosis in ICC via Src-mediated post-translational modifications. Src phosphorylation of USP10 stabilizes FSP1 by removing K48-linked polyubiquitin. Src activates NMT1 to enhance FSP1 myristoylation and membrane localization.
Keywords: MUC1; ferroptosis; intrahepatic cholangiocarcinoma; post‐translational modification.
© 2025 The Author(s). Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
