The genetic influence of sex on gene expression for blood in pigs
- PMID: 41068573
- PMCID: PMC12512681
- DOI: 10.1186/s12864-025-12029-3
The genetic influence of sex on gene expression for blood in pigs
Abstract
Background: Pigs are one of the most important farm animals in the agrifood industry. Many complex traits and patterns of gene expression exhibit sexual dimorphisms in pigs. However, the impact of sex on gene expression remains poorly understood.
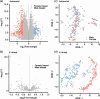
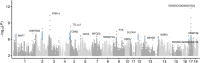
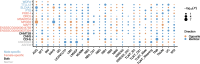
Results: In this study, we utilized the gene expression data of blood tissue derived from PigGTEx project to explore the genetic influence of sex on gene expression in pigs. Differential gene expression analysis identified 116 male-biased and 248 female-biased genes. Sex-combined and sex-stratified cis-heritability (cis-h2) were highly positively correlated, while the low correlation were observed between male-stratified and female-stratified cis-h2. Sex-interaction expression quantitative trait locus (eQTL) mapping identified 16 genes with at least one sex-biased eQTL (sb-eGenes) in blood, including 7 female-specific eGenes and 4 male-specific eGenes. Notable examples included the immunology-associated male-specific eGene SLC4A1 and the female-specific eGene PRR14, illustrating sex-specific regulation of gene expression in blood. We further found that sb-eGenes were associated with various complex traits through distinct genetic regulation mechanisms. For example, the male-specific gene SLC4A1 was associated with average daily gain with the identical effect, while the female-specific gene MFGE8 exhibited opposite effect.
Conclusions: This study revealed sex-biased gene expression patterns and sex-dependent regulatory effect of gene expression of blood tissues in pigs. Additionally, this study found the sexually dimorphic regulation of gene expression underlying complex traits. These findings provided a comprehensive insight and resource and advance our understanding of sexual dimorphism in genetic mechanism underlying complex traits in blood.
Keywords: Blood; Expression quantitative trait locus; Gene expression; Pig; Sex-biased gene.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not appliable. Consent for publication: Not appliable. Competing interests: The authors declare no competing interests.
Figures

References
-
- Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16:197–212. 10.1038/nrg3891. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

