A methylome-wide association study of major depression with out-of-sample case-control classification and trans-ancestry comparison
- PMID: 41069367
- PMCID: PMC12504109
- DOI: 10.1038/s44220-025-00486-4
A methylome-wide association study of major depression with out-of-sample case-control classification and trans-ancestry comparison
Abstract
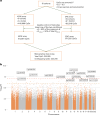
Major depression (MD) is a leading cause of global disease burden, and both experimental and population-based studies suggest that differences in DNA methylation may be associated with the condition. However, previous DNA methylation studies have, so far, not been widely replicated, suggesting a need for larger meta-analysis studies. Here we conducted a meta-analysis of methylome-wide association analysis for lifetime MD across 18 studies of 24,754 European-ancestry participants (5,443 MD cases) and an East Asian sample (243 cases, 1,846 controls). We identified 15 CpG sites associated with lifetime MD with methylome-wide significance. The methylation score created using the methylome-wide association analysis summary statistics was significantly associated with MD status in an out-of-sample classification analysis (area under the curve 0.53). Methylation score was also associated with five inflammatory markers, with the strongest association found with tumor necrosis factor beta. Mendelian randomization analysis revealed 23 CpG sites potentially causally linked to MD, with 7 replicated in an independent dataset. Our study provides evidence that variations in DNA methylation are associated with MD, and further evidence supporting involvement of the immune system.
Keywords: DNA methylation; Depression; Epigenomics; Predictive markers.
© The Author(s) 2025.
Conflict of interest statement
Competing interestsQ.S.L. is an employee of Janssen Research & Development, LLC. Q.S.L. owns stock and/or stock options in Johnson & Johnson. H.J.G. has received travel grants and speakers’ honoraria from Indorsia, Neuraxpharm, Servier and Janssen Cilag. T. Kircher received unrestricted educational grants from Servier, Janssen, Recordati, Aristo, Otsuka, neuraxpharm. The remaining authors declare no competing interests.
Figures



References
-
- Sullivan, P. F., Neile, M. C. & Kendler, K. S. Genetic epidemiology of major depression: review and meta-analysis. Am. J. Psychiatry157, 1552–1562 (2000). - PubMed
LinkOut - more resources
Full Text Sources
