Glioma‑associated microglia and macrophages as a potential target for mTOR inhibition in glioblastoma
- PMID: 41070610
- PMCID: PMC12529088
- DOI: 10.3892/mmr.2025.13708
Glioma‑associated microglia and macrophages as a potential target for mTOR inhibition in glioblastoma
Abstract
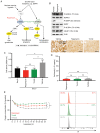
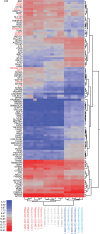
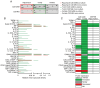
Glioma‑associated microglia/macrophages (GAM) constitute the predominant immune cell population in glioblastoma (GB). Both GB cells and GAM exhibit upregulated mTOR signaling. The present study aimed to investigate the effects of pharmacological mTOR inhibition (mTORi) specifically on GAM. The effects of mTORi on signal transduction, cell growth and viability were analyzed in immortalized microglia cell lines. Additionally, a comprehensive analysis of the GAM phenotype was conducted, including whole transcriptome analyses and cytokine profiling. Effects were investigated in a tumor cell/GAM co‑culture model under mTORi with rapamycin or torin2 or treatment with temozolomide, the standard chemotherapy agent for patients with GB. In the in vitro model, mTORi had significant effects on central biological functions of GAM, resulting in reduced proliferation and oxygen consumption. Additionally, treatment with mTORi induced a pro‑inflammatory phenotype in microglia cell lines. These findings demonstrate the relevance of mTOR signaling on GAM biology. Moreover, they provide rationales for therapeutic interventions targeting mTOR signaling specifically in GAM as a potential novel treatment strategy.
Keywords: glioma; mTOR; microglia; rapamycin; tumor microenvironment.
Conflict of interest statement
JPS has received honoraria for lectures, advisory board participation, consulting or travel grants from Abbvie, Roche, Boehringer, Bristol-Myers Squibb, Medac, Mundipharma, Servier and UCB. MHM is meanwhile an employee of Sanofi. MWR has received a research grant from UCB as well as honoraria for advisory board participation from Alexion and Servier. PSZ, MS, JS, NIL, JBW, BS, BR, KJW, ALL, AB, KHP, LS and PNH report no disclosures relevant to the manuscript.
Figures


References
-
- Mandel JJ, Yust-Katz S, Patel AJ, Cachia D, Liu D, Park M, Yuan Y, Kent TA, de Groot JF. Inability of positive phase II clinical trials of investigational treatments to subsequently predict positive phase III clinical trials in glioblastoma. Neuro Oncol. 2018;20:113–122. doi: 10.1093/neuonc/nox144. - DOI - PMC - PubMed
-
- Bowman RL, Klemm F, Akkari L, Pyonteck SM, Sevenich L, Quail DF, Dhara S, Simpson K, Gardner EE, Iacobuzio-Donahue CA, et al. Macrophage ontogeny underlies differences in tumor-specific education in brain malignancies. Cell Rep. 2016;17:2445–2459. doi: 10.1016/j.celrep.2016.10.052. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

