Emerging insights into alternative end‑joining: Mechanisms, genome instability and therapeutic opportunities in cancer (Review)
- PMID: 41070620
- PMCID: PMC12543316
- DOI: 10.3892/ijo.2025.5809
Emerging insights into alternative end‑joining: Mechanisms, genome instability and therapeutic opportunities in cancer (Review)
Abstract
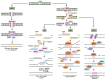
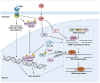
Genome instability is a central hallmark of cancer, driven by aberrant DNA damage responses that facilitate tumor evolution and resistance to therapy. Although canonical non‑homologous end joining and homologous recombination are well‑characterized pathways for repairing DNA double‑strand breaks (DSBs), recent advances have revealed that cancer cells increasingly depend on alternative end‑joining (alt‑EJ) to survive persistent DNA damage that arises from intrinsic stresses or external therapies. Alt‑EJ, characterized by its reliance on microhomologous sequences at DSB sites, promotes mutation accumulation and chromosomal rearrangements, thereby driving genomic instability and tumor progression. Despite its pivotal role in cancer biology, the molecular regulation, contextual determinants and dualistic role of alt‑EJ in maintaining genome integrity compared with promoting instability remain incompletely understood. The present review integrated the latest mechanistic insights into alt‑EJ, elucidated its regulatory networks and interactions with canonical DSB repair pathways and discussed its consequences for cancer genome integrity and evolution. Furthermore, it highlighted the emerging potential of alt‑EJ as a therapeutic vulnerability for cancer, underscoring the urgent need to translate these discoveries into innovative treatment strategies aimed at overcoming therapy resistance and improving patient outcomes.
Keywords: DNA damage response; DNA double strand break repair; alternative end‑joining; cancer therapeutic targets; genome instability.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

