Functional recovery of islet β cells in human type 2 diabetes: Transcriptome signatures unveil therapeutic approaches
- PMID: 41071888
- PMCID: PMC12513425
- DOI: 10.1126/sciadv.ads2905
Functional recovery of islet β cells in human type 2 diabetes: Transcriptome signatures unveil therapeutic approaches
Abstract
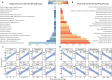
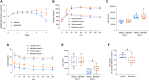
Remission of type 2 diabetes (T2D) can occur after hypocaloric diet, bariatric surgery, or pharmacological treatments and associates with improved β cell function. Here, we studied islets from nondiabetic (n = 15) and T2D (n = 21) donors. We examined whether T2D β cell dysfunction can be rescued, charted the underlying molecular mechanisms by RNA sequencing, and mined transcriptomes for drug targets. Glucose responsiveness of T2D β cells improved in 60% of preparations after 3-day culture in euglycemic conditions. This was accompanied by changes in expression of >400 genes involved in functional or inflammatory pathways. Drug repurposing and target identification analyses predicted chemical and genetic hits, including JAK inhibitors, which were validated in a β cell line, human islets, and db/db mice. Therefore, defective β cell glucose responsiveness in T2D can recover, demonstrating β cell functional plasticity. The recovery associates with transcriptomic traits, pointing to targetable defects to induce T2D remission.
Figures





References
-
- Ahmad E., Lim S., Lamptey R., Webb D. R., Davies M. J., Type 2 diabetes. Lancet 400, 1803–1820 (2022). - PubMed
-
- Alonso L., Piron A., Morán I., Guindo-Martínez M., Bonàs-Guarch S., Atla G., Miguel-Escalada I., Royo R., Puiggròs M., Garcia-Hurtado X., Suleiman M., Marselli L., Esguerra J. L. S., Turatsinze J.-V., Torres J. M., Nylander V., Chen J., Eliasson L., Defrance M., Amela R., MAGIC, Mulder H., Gloyn A. L., Groop L., Marchetti P., Eizirik D. L., Ferrer J., Mercader J. M., Cnop M., Torrents D., TIGER: The gene expression regulatory variation landscape of human pancreatic islets. Cell Rep. 37, 109807 (2021). - PMC - PubMed
-
- Cnop M., Abdulkarim B., Bottu G., Cunha D. A., Igoillo-Esteve M., Masini M., Turatsinze J.-V., Griebel T., Villate O., Santin I., Bugliani M., Ladriere L., Marselli L., McCarthy M. I., Marchetti P., Sammeth M., Eizirik D. L., RNA sequencing identifies dysregulation of the human pancreatic islet transcriptome by the saturated fatty acid palmitate. Diabetes 63, 1978–1993 (2014). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

