SETBP1 variants outside the degron disrupt DNA-binding, transcription and neuronal differentiation capacity to cause a heterogeneous neurodevelopmental disorder
- PMID: 41073373
- PMCID: PMC12514306
- DOI: 10.1038/s41467-025-64074-x
SETBP1 variants outside the degron disrupt DNA-binding, transcription and neuronal differentiation capacity to cause a heterogeneous neurodevelopmental disorder
Abstract
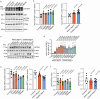
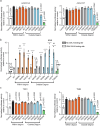
Different types of germline de novo SETBP1 variants cause clinically distinct and heterogeneous neurodevelopmental disorders: Schinzel-Giedion syndrome (SGS, via missense variants at a critical degron region) and SETBP1-haploinsufficiency disorder. However, due to the lack of systematic investigation of genotype-phenotype associations of different types of SETBP1 variants, and limited understanding of its roles in neurodevelopment, the extent of clinical heterogeneity and how this relates to underlying pathophysiological mechanisms remains elusive. This imposes challenges for diagnosis. Here, we present a comprehensive investigation of the largest cohort to date of individuals carrying SETBP1 missense variants outside the degron region (n = 18). We performed thorough clinical and speech phenotyping with functional follow-up using cellular assays and transcriptomics. Our findings suggest that such variants cause a clinically and functionally variable developmental syndrome, showing only partial overlaps with classical SGS and SETBP1-haploinsufficiency disorder. We provide evidence of loss-of-function pathophysiological mechanisms impairing ubiquitination, DNA-binding, transcription, and neuronal differentiation capacity and morphologies. In contrast to SGS and SETBP1 haploinsufficiency, these effects are independent of protein abundance. Overall, our study provides important novel insights into diagnosis, patient care, and aetiology of SETBP1-related disorders.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures






References
-
- Hoischen, A. et al. De novo mutations of SETBP1 cause Schinzel–Giedion syndrome. Nat. Genet.42, 483–485 (2010). - PubMed
-
- Schinzel, A. & Giedion, A. A syndrome of severe midface retraction, multiple skull anomalies, clubfeet, and cardiac and renal malformations in sibs. Am. J. Med. Genet.1, 361–375 (1978). - PubMed
-
- Minn, D. et al. Further clinical and sensorial delineation of Schinzel–Giedion syndrome: report of two cases. Am. J. Med. Genet.109, 211–217 (2002). - PubMed
-
- AL-Mudaffer, M. et al. Clinical and radiological findings in Schinzel–Giedion syndrome. Eur. J. Pediatr.167, 1399–1407 (2008). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

