Estrogen-related receptor gene expression associates with sex differences in cortical atrophy in isolated REM sleep behavior disorder
- PMID: 41073384
- PMCID: PMC12514017
- DOI: 10.1038/s41467-025-63829-w
Estrogen-related receptor gene expression associates with sex differences in cortical atrophy in isolated REM sleep behavior disorder
Abstract
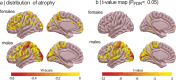
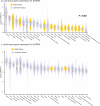
Isolated REM sleep behavior disorder, characterized by dream-enacting movements during REM sleep, is a male-predominant parasomnia and the strongest prodromal marker of synucleinopathies. Individuals with this disorder show cortical atrophy whose regional distribution covaries with gene expression patterns measured in the healthy human brain. However, the effect of sex on these brain changes remains unknown. The study objective is to comprehensively assess sex differences in cortical morphology and to characterize the healthy-brain gene expression correlates of brain abnormalities using the largest international multicentric MRI dataset of polysomnography-confirmed patients. Males have significantly more extensive and severe cortical thinning compared to females, despite similar age and clinical features. Imaging transcriptomics analyses indicate that regions affected in female patients map onto areas with higher expression of estrogen-related receptor genes, particularly ESRRG and ESRRA, in the healthy brain. These findings support potential sex-specific neuroprotection in the prodromal stages of synucleinopathies and may inform personalized and targeted therapeutic strategies.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: There are no competing interests.
Figures


References
-
- American Academy of Sleep Medicine. The International Classification of Sleep Disorders — Third Edition (ICSD-3). (American Academy of Sleep Medicine, 2014).
MeSH terms
Substances
Grants and funding
- NU21-04-00535/Agentura Pro Zdravotnický Výzkum České Republiky (Czech Health Research Council)
- J-2101/PUK_/Parkinson's UK/United Kingdom
- J-2101/PUK_/Parkinson's UK/United Kingdom
- #1095127/Department of Health | National Health and Medical Research Council (NHMRC)
- PPG-2023-0000000122/Parkinson Canada
LinkOut - more resources
Full Text Sources

