A microfluidic platform for the co-culturing of microtissues with continuously recirculating suspension cells
- PMID: 41073406
- PMCID: PMC12514218
- DOI: 10.1038/s41378-025-01028-9
A microfluidic platform for the co-culturing of microtissues with continuously recirculating suspension cells
Abstract
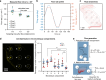
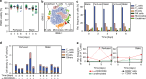
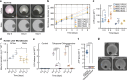
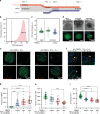
In vitro evaluation of novel therapeutic approaches often fails to reliably predict efficacy and toxicity, especially when recapitulating conditions involving recirculating cells. Current testing strategies are often based on static co-culturing of cells in suspension and 3D tissue models, where cell sedimentation on the target tissue can occur. The observed effects may then mostly be a consequence of sedimentation and of the corresponding forced cell-tissue interactions. The realization of continuous medium flow helps to better recapitulate physiological conditions and cell-tissue interactions. To tackle current limitations of perfused organ-on-chip approaches, we developed a microfluidic chip and operation concept, which prevents undesired sedimentation and accumulation of suspended cells during multiple days by relying on gravity-driven perfusion. Our platform, which we termed "human immune flow (hiFlow) chip", enables to co-culture cells in suspension with up to 7 preformed microtissue models. Here, we present the design principle and operation of the platform, and we validate its performance by culturing cells and microtissues of a variety of different origins. Cells and tissues could be monitored on chip via high-resolution microscopy, while cell suspensions and microtissues could be easily retrieved for off-chip analysis. Our results demonstrate that primary immune cells and a range of different spheroid models of healthy and diseased tissues can be maintained for over 6 days on chip. As proof-of-concept cell-tissue interaction assay, we used an antibody treatment against diffuse midline glioma, a highly aggressive pediatric tumor. We are confident that our platform will help to increase the prediction power of in vitro preclinical testing of novel therapeutics that rely on the interaction of circulating cells with organ tissues.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: O.F. is part of the management team of InSphero AG, and T.H., L.H., L.L., and M.R. are employees of InSphero AG, which commercializes the microfluidic culturing device.
Figures

References
-
- Kang, S. et al. Complex in vitro models positioned for impact to drug testing in pharma: a review. Biofabrication 16 (2024). - PubMed
-
- Leung, C. M. et al. A guide to the organ-on-a-chip. Nat. Rev. Methods Prim2, 33 (2022).
LinkOut - more resources
Full Text Sources
Research Materials
