Integrative functional genomics analysis of Kaposi sarcoma cohorts
- PMID: 41073438
- PMCID: PMC12514283
- DOI: 10.1038/s41598-025-08564-4
Integrative functional genomics analysis of Kaposi sarcoma cohorts
Abstract
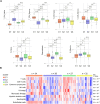
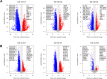
Kaposi sarcoma (KS) is an AIDS-defining cancer and a significant global health challenge caused by KS-associated herpesvirus (KSHV). NGS-based approaches have profiled KS lesions in a minimal number of studies compared with other neoplastic diseases. Here we present a compiled and harmonized dataset of 131 KS and non-tumor cutaneous samples in the context of their predicted pathway activities, immune infiltrate, KSHV and HIV gene expression profiles, and their associated clinical data representing patient populations from Argentina, United States (USA), and Sub-Saharan Africa cohorts. RNA-seq data from 9 Argentinian KS lesions were generated and integrated with previously published datasets derived from the USA and sub-Saharan African cohorts from Tanzania, Zambia, and Uganda. An unsupervised analysis of 131 KS-related samples allowed us to identify four KS clusters based on their host and KSHV gene expression profiles, immune infiltrate, and the activity of specific signaling pathways. The compiled RNA-seq profile is shared with the research community through the UCSC Xena browser for further visualization, download, and analysis ( https://kaposi.xenahubs.net/ ). These resources will allow biologists without bioinformatics knowledge to explore and correlate the host and viral transcriptome in a curated dataset of different KS RNA-seq-based cohorts, which can lead to novel biological insights and biomarker discovery.
Keywords: Kaposi’s sarcoma; RNAseq; Transcriptome; Xena browser.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics declarations: MJG is a consultant for the Fred Hutchinson Cancer Research Center. The other authors declare no competing interests. Competing interests: The authors declare no competing interests.
Figures



Update of
-
Integrative Functional Genomics Analysis of Kaposi Sarcoma Cohorts.Res Sq [Preprint]. 2025 Mar 11:rs.3.rs-6146471. doi: 10.21203/rs.3.rs-6146471/v1. Res Sq. 2025. Update in: Sci Rep. 2025 Oct 10;15(1):35413. doi: 10.1038/s41598-025-08564-4. PMID: 40162228 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

