Population-scale gene-based analysis of whole-genome sequencing provides insights into metabolic health
- PMID: 41073786
- PMCID: PMC12513836
- DOI: 10.1038/s41588-025-02364-2
Population-scale gene-based analysis of whole-genome sequencing provides insights into metabolic health
Abstract
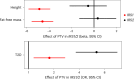
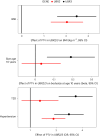
In addition to its coverage of the noncoding genome, whole-genome sequencing (WGS) may better capture the coding genome than exome sequencing. Here we sought to exploit this and identify new rare, protein-coding variants associated with metabolic health in WGS data (n = 708,956) from the UK Biobank and All of Us studies. Identified genes highlight new biological mechanisms, including protein-truncating variants (PTVs) in the DNA double-strand break repair gene RIF1 that have a substantial effect on body mass index (2.66 kg m-2, s.e. 0.43, P = 3.7 × 10-10). UBR3 is an intriguing example where PTVs independently increase body mass index and type 2 diabetes risk. Furthermore, PTVs in IRS2 have a substantial effect on type 2 diabetes (odds ratio 6.4 (3.7-11.3), P = 9.9 × 10-14, 34% case prevalence among carriers) and were also associated with chronic kidney disease independent of diabetes status, suggesting an important role for IRS2 in maintaining renal health. Our study demonstrates that large-scale WGS provides new mechanistic insights into human metabolic phenotypes through improved capture of coding sequences.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.L., A.C., Y.L., J.D., C.B.-D. and R.S. are employees and stockholders of GSK. J.R.B.P. and E.J.G. are employees and shareholders of Insmed. J.R.B.P. receives research funding from GSK. Y.Z. was a UK University worker at GSK during this work. S.O.ʼR. has undertaken remunerated consultancy work for Pfizer, Third Rock Ventures, AstraZeneca, NorthSea Therapeutics and Courage Therapeutics and is a scientific founder of Marea Therapeutics. S.L. performs paid consultancy for Eolas Medical. The other authors declare no competing interests.
Figures


References
-
- Uffelmann, E. et al. Genome-wide association studies. Nat. Rev. Methods Prim.1, 59 (2021). - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

