Segmenting beyond the imaging data: creation of anatomically valid edentulous mandibular geometries for surgical planning using artificial intelligence
- PMID: 41074944
- PMCID: PMC12515113
- DOI: 10.1007/s00784-025-06471-6
Segmenting beyond the imaging data: creation of anatomically valid edentulous mandibular geometries for surgical planning using artificial intelligence
Abstract
Background and objectives: Mandibular reconstruction following continuity resection due to tumor ablation or osteonecrosis remains a significant challenge in maxillofacial surgery. Virtual surgical planning (VSP) relies on accurate segmentation of the mandible, yet existing AI models typically include teeth, making them unsuitable for planning of autologous transplants dimensions aiming for reconstructing edentulous mandibles optimized for dental implant insertion. This study investigates the feasibility of using deep learning-based segmentation to generate anatomically valid, toothless mandibles from dentate CT scans, ensuring geometric accuracy for reconstructive planning.
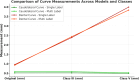
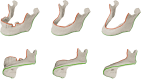
Methods: A two-stage convolutional neural network (CNN) approach was employed to segment mandibles from computed tomography (CT) data. The dataset (n = 246) included dentate, partially dentate, and edentulous mandibles. Ground truth segmentations were manually modified to create Class III (moderate alveolar atrophy) and Class V (severe atrophy) models, representing different degrees of post-extraction bone resorption. The AI models were trained on the original (O), Class III (Cl. III), and Class V (Cl. V) datasets, and performance was evaluated using Dice similarity coefficients (DSC), average surface distance, and automatically detected anatomical curvatures.
Results: AI-generated segmentations demonstrated high anatomical accuracy across all models, with mean DSCs exceeding 0.94. Accuracy was highest in edentulous mandibles (DSC 0.96 ± 0.014) and slightly lower in fully dentate cases, particularly for Class V modifications (DSC 0.936 ± 0.030). The caudolateral curve remained consistent, confirming that baseline mandibular geometry was preserved despite alveolar ridge modifications.
Conclusions: This study confirms that AI-driven segmentation can generate anatomically valid edentulous mandibles from dentate CT scans with high accuracy. The innovation of the work is the precise adaptation of alveolar ridge geometry, making it a valuable tool for patient-specific virtual surgical planning in mandibular reconstruction.
Keywords: Computed tomography; Convolutional neural networks; Deep learning; Mandible; Medical image analysis; Reconstruction; Segmentation; Virtual surgical planning.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: S.R. reports a relationship with Inzipio GmbH that includes: co-founder, employment and shares. T. P. reports a relationship with Inzipio GmbH that includes: co-founder, employment and shares. A. M. reports a relationship with Inzipio GmbH that includes: co-founder and shares. Other authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Ethical approval: Institutional approval (EK 260/20) of the local ethics committee of RWTH Aachen University Hospital was obtained. Informed consent: Due to the retrospective nature of the study, the Independent Ethics Committee of the Faculty of Medicine of RWTH Aachen University Hospital waived the need to obtain informed consent.
Figures





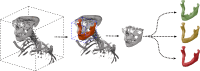

References
-
- 1. Mücke T, Hölzle F, Loeffelbein DJ, Ljubic A, Kesting M, Wolff K-D, Mitchell DA (2011) Maxillary reconstruction using microvascular free flaps. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 111:51–7. 10.1016/j.tripleo.2010.03.042 - PubMed
-
- 2. Brown J, Bekiroglu F, Shaw R (2010) Indications for the scapular flap in reconstructions of the head and neck. Br J Oral Maxillofac Surg 48:331–7. 10.1016/j.bjoms.2009.09.013 - PubMed
-
- 3. Jones NF, Swartz WM, Mears DC, Jupiter JB, Grossman A (1988) The “double barrel” free vascularized fibular bone graft. Plast Reconstr Surg 81:378–385 - PubMed
-
- 4. Pucci R, Weyh A, Smotherman C, Valentini V, Bunnell A, Fernandes R (2020) Accuracy of virtual planned surgery versus conventional free-hand surgery for reconstruction of the mandible with osteocutaneous free flaps. Int J Oral Maxillofac Surg 49:1153–1161. 10.1016/j.ijom.2020.02.018 - PubMed
-
- 5. Gillingham RL, Mutsvangwa TEM, van der Merwe J (2023) Reconstruction of the mandible from partial inputs for virtual surgery planning. Med Eng Phys 111. 10.1016/j.medengphy.2022.103934 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

