PDLIM4 drives gastric cancer malignant progression and cisplatin resistance by inhibiting HSP70 ubiquitination and degradation via competitive interaction with STUB1
- PMID: 41076533
- PMCID: PMC12514821
- DOI: 10.1186/s12951-025-03720-4
PDLIM4 drives gastric cancer malignant progression and cisplatin resistance by inhibiting HSP70 ubiquitination and degradation via competitive interaction with STUB1
Abstract
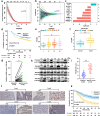
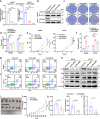
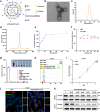
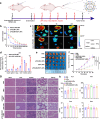
Gastric cancer (GC) frequently shows malignant progression and resistance to chemotherapy due to complex molecular regulatory processes, leading to a poor prognosis. Our study elucidates that PDZ and LIM domain protein 4 (PDLIM4) are highly expressed in GC, thereby promoting the malignant progression and cisplatin (DDP) resistance of GC. Mechanistically, PDLIM4, via its C-terminal and intermediate regions, impedes the ubiquitination and proteasomal degradation of Heat Shock Protein 70 (HSP70) by competing with the STIP1 homology and U-box containing protein 1 (STUB1), subsequently activating the MAPK signaling pathway. In addition, we synthesized lipid nanoparticles (LNPs), including siPDLIM4 LNPs, DDP LNPs, and siPDLIM4/DDP LNPs. Experiments further indicated that siPDLIM4 LNPs and DDP LNPs have an anti-tumor property, while siPDLIM4/DDP LNPs exhibit the most significant anti-tumor efficacy. In summary, our research identifies PDLIM4 as a facilitator of malignant progression and DDP resistance in GC cells, targeting PDLIM4 not only inhibits the malignant progression of GC, but also provides an effective strategy to enhance DDP sensitivity in GC treatment, emphasizing its potential as a therapeutic target.
Keywords: DDP; Gastric cancer; Lipid nanoparticles; PDLIM4; STUB1/HSP70; Ubiquitination.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The animal research received approval from the Ethics Committee of the First Affiliated Hospital of Nanchang University (Ethics Number: CDYFY-IACUC-202309QR019). The Ethics Committee of the First Affiliated Hospital of Nanchang University approved the study, and all participants provided informed consent (Approval Number: (2025) CDYFYYLK (04–030)). Consent for publication: All authors have read and approved the final manuscript. Competing interests: The authors declare no competing interests.
Figures






References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca-a Cancer Journal for Clinicians. 2024;74:229–63. - PubMed
-
- Sundar R, Nakayama I, Markar SR, Shitara K, van Laarhoven HWM, Janjigian YY, et al. Gastric cancer. Lancet (London, England). 2025. 10.1016/S0140-6736(25)00052-2. - PubMed
-
- Cho HJ, Baek KE, Park S-M, Kim I-K, Nam I-K, Choi Y-L, et al. RhoGDI2 confers gastric cancer cells resistance against cisplatin-induced apoptosis by upregulation of Bcl-2 expression. Cancer Lett. 2011;311:48–56. - PubMed
MeSH terms
Substances
Grants and funding
- 20224BAB216011/Natural Science Foundation of Jiangxi Province
- 20213AAG01013/the major science and technology R & D projects of Jiangxi Province
- 82472225/the National Nature Science Foundation in China (NSFC)
- jxsq2023201037/Jiangxi Province Science and Technology Innovation Talent Project
- 2024SSY06171/Science and technology innovation base plan of Jiangxi Province-Key Laboratory of Jiangxi Province for Transfusion Medicine
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

